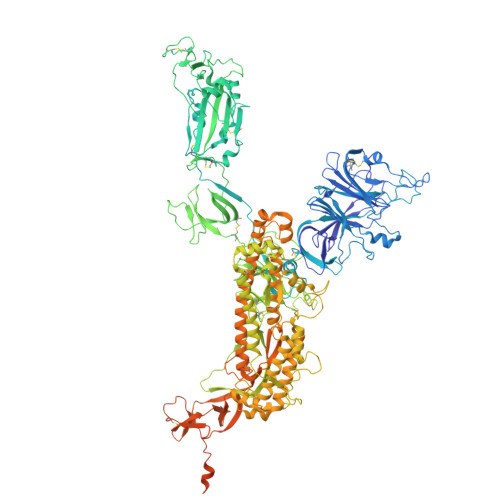
Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
Chang, Y.H., Hsu, M.F., Chen, W.N., Wu, M.H., Kong, W.L., Lu, M.J., Huang, C.H., Chang, F.J., Chang, L.Y., Tsai, H.Y., Tung, C.P., Yu, J.H., Kuo, Y., Chou, Y.C., Bai, L.Y., Chang, Y.C., Chen, A.Y., Chen, C.C., Chen, Y.H., Liao, C.C., Chang, C.S., Liang, J.J., Lin, Y.L., Angata, T., Hsu, S.D., Lin, K.I.(2024) JCI Insight 9
- PubMed: 38775156
- DOI: https://doi.org/10.1172/jci.insight.179726
- Primary Citation of Related Structures:
8XAL, 8XBF - PubMed Abstract:
Since its emergence, SARS-CoV-2 has been continuously evolving, hampering the effectiveness of current vaccines against COVID-19. mAbs can be used to treat patients at risk of severe COVID-19. Thus, the development of broadly protective mAbs and an understanding of the underlying protective mechanisms are of great importance. Here, we isolated mAbs from donors with breakthrough infection with Omicron subvariants using a single-B cell screening platform. We identified a mAb, O5C2, which possesses broad-spectrum neutralization and antibody-dependent cell-mediated cytotoxic activities against SARS-CoV-2 variants, including EG.5.1. Single-particle analysis by cryo-electron microscopy revealed that O5C2 targeted an unusually large epitope within the receptor-binding domain of spike protein that overlapped with the angiotensin-converting enzyme 2 binding interface. Furthermore, O5C2 effectively protected against BA.5 Omicron infection in vivo by mediating changes in transcriptomes enriched in genes involved in apoptosis and interferon responses. Our findings provide insights into the development of pan-protective mAbs against SARS-CoV-2.
- Genomics Research Center, Academia Sinica, Taipei, Taiwan.
Organizational Affiliation: