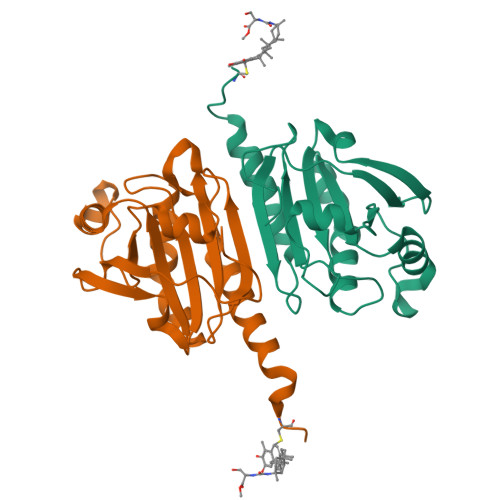
Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
Wang, J., Rong, Q., Ye, L., Fang, B., Zhao, Y., Sun, Y., Zhou, H., Wang, D., He, J., Cui, Z., Zhang, Q., Kang, D., Hu, L.(2024) J Med Chem 67: 7197-7223
- PubMed: 38655686
- DOI: https://doi.org/10.1021/acs.jmedchem.4c00051
- Primary Citation of Related Structures:
8X71, 8X73 - PubMed Abstract:
Fms-like tyrosine receptor kinase 3 (FLT3) proteolysis-targeting chimeras (PROTACs) represent a promising approach to eliminate the resistance of FLT3 inhibitors. However, due to the poor druggability of PROTACs, the development of orally bioavailable FLT3-PROTACs faces great challenges. Herein, a novel orally bioavailable FLT3-ITD degrader A20 with excellent pharmacokinetic properties was discovered through reasonable design. A20 selectively inhibited the proliferation of FLT3-ITD mutant acute myeloid leukemia (AML) cells and potently induced FLT3-ITD degradation through the ubiquitin-proteasome system. Notably, oral administration of A20 resulted in complete tumor regression on subcutaneous AML xenograft models. Furthermore, on systemic AML xenograft models, A20 could completely eliminate the CD45 + CD33 + human leukemic cells in murine and significantly prolonged the survival time of mice. Most importantly, A20 exerted significantly improved antiproliferative activity against drug-resistant AML cells compared to existing FLT3 inhibitors. These findings suggested that A20 could serve as a promising drug candidate for relapsed or refractory AML.
Organizational Affiliation:
Jiangsu Key Laboratory for Functional Substance of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing 210023, P. R. China.