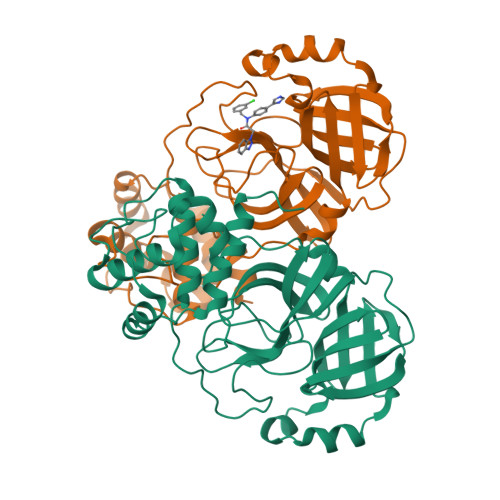
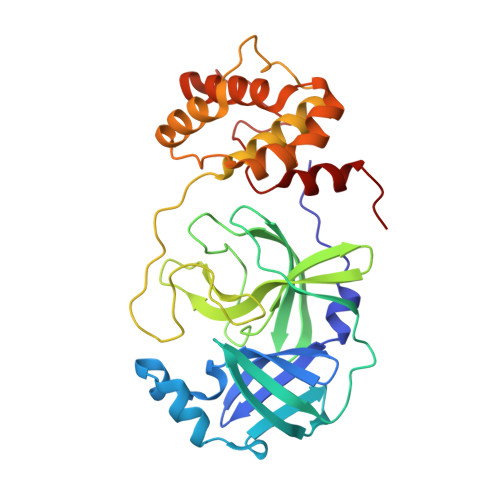
Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Jiang, H., Zou, X., Zhou, X., Zhang, J., Li, J.(2024) Biochem Biophys Res Commun 692: 149352-149352
- PubMed: 38056159
- DOI: https://doi.org/10.1016/j.bbrc.2023.149352
- Primary Citation of Related Structures:
8WZP, 8WZQ - PubMed Abstract:
SARS-CoV-2 constantly circulates and evolves worldwide, generating many variants and posing a menace to global health. It is urgently needed to discover effective medicines to treat the disease caused by SARS-CoV-2 and its variants. An established target for anti-SARS-CoV-2 drug discovery is the main protease (M pro ), since it exerts an irreplaceable action in viral life cycle. CCF0058981, derived from ML300, is a non-covalent inhibitor that exhibits low nanomolar potency against SARS-CoV-2 M pro and submicromolar anti-SARS-CoV-2 activity, thereby providing a valuable starting point for drug design. However, structural basis underlying inhibition of SARS-CoV-2 M pro by CCF0058981 remains undetermined. In this study, the crystal structures of CCF0058981 in complex with two SARS-CoV-2 M pro mutants (M49I and V186F), which have been identified in the recently emerged Omicron subvariants, were solved. Structural analysis defined the pivotal molecular factors responsible for the interactions between CCF0058981 and these two M pro mutants, and revealed the binding modes of CCF0058981 to M pro M49I and V186F mutants. These data not only provide structural insights for SARS-CoV-2 M pro inhibition by CCF0058981, but also add to develop effective broad-spectrum drugs against SARS-CoV-2 as well as its variants.
Organizational Affiliation:
School of Basic Medical Sciences, Nanchang University, Nanchang, 330031, China.