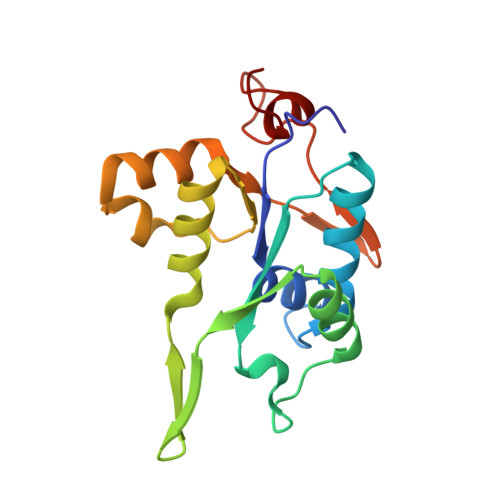
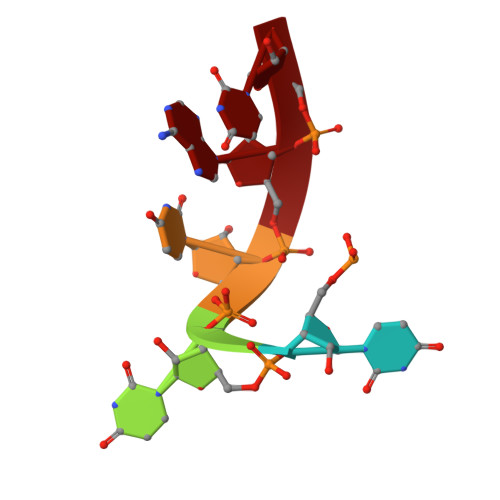
Crystal structure of NYN domain of Human KHNYN in complex with single strand RNA.
Hong, S., Choe, J.(2024) Biochem Biophys Res Commun 738: 150545-150545
- PubMed: 39167961
- DOI: https://doi.org/10.1016/j.bbrc.2024.150545
- Primary Citation of Related Structures:
8WZC - PubMed Abstract:
KHNYN protein with a KH-like domain and a NYN endoribonuclease domain interacts with Zinc-finger antiviral protein (ZAP). ZAP isoforms recognize viral or cellular RNAs and recruit KHNYN to form the ZAP: KHNYN complex. Although the structures of several PIN/NYN domains have been determined, the precise substrate RNA binding mode remains poorly understood. This study presents the crystal structure of a complex of the NYN domain of KHNYN and a 7mer RNA from interferon lambda3 (IFNL3). Our structural analysis reveals that NYN domain of human KHNYN shares structural similarities with other NYN domains of ZC3H12Ã C proteins. The RNA is bound in the central groove region of the protein, facilitated by interactions including coordination by two Mg 2+ ions, hydrophobic interactions, and hydrogen bonds. In the observed RNA-protein complex, the U 5 , A 6 , and U 7 bases are stacked on top of one another, while U 3 and U 4 bases adopt an "open" conformation (as opposed to base-stacked), forming a U-shaped overall structure. Mutagenesis studies underscore the significance of residues involved in RNA binding for RNase activity. Interestingly, NYN domain of human KHNYN forms a head-to-tail dimer in the crystal, a structural feature also observed in other homologous PIN/NYN proteins, with a residue from the symmetry mate contributing to hydrophobic interactions with the bound RNA.
Organizational Affiliation:
Department of Life Science, University of Seoul, Seoul, 02504, Republic of Korea.