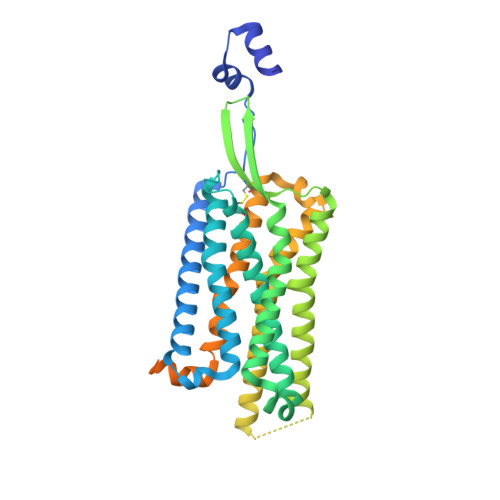
Structural basis for recognition of 26RFa by the pyroglutamylated RFamide peptide receptor.
Jin, S., Guo, S., Xu, Y., Li, X., Wu, C., He, X., Pan, B., Xin, W., Zhang, H., Hu, W., Yin, Y., Zhang, T., Wu, K., Yuan, Q., Xu, H.E., Xie, X., Jiang, Y.(2024) Cell Discov 10: 58-58
- PubMed: 38830850
- DOI: https://doi.org/10.1038/s41421-024-00670-3
- Primary Citation of Related Structures:
8WZ2 - PubMed Abstract:
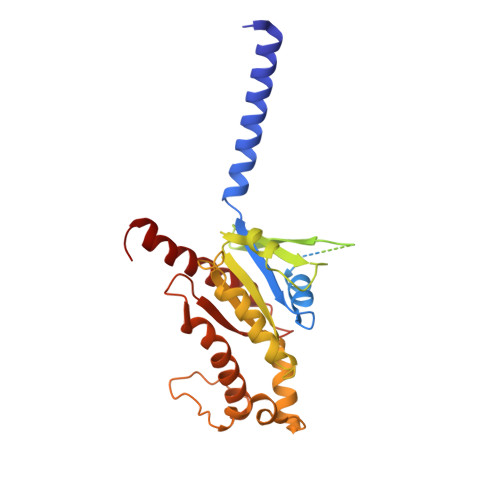
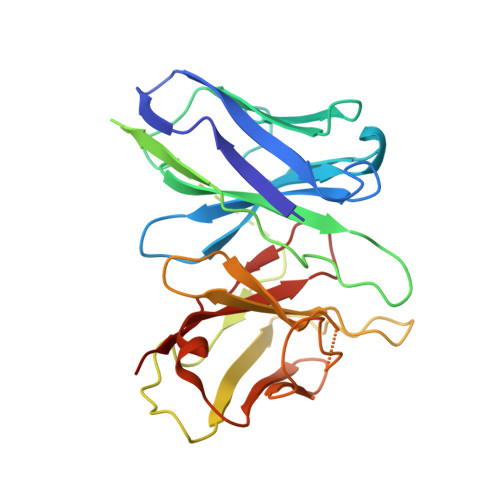
The neuropeptide 26RFa, a member of the RF-amide peptide family, activates the pyroglutamylated RF-amide peptide receptor (QRFPR), a class A GPCR. The 26RFa/QRFPR system plays critical roles in energy homeostasis, making QRFPR an attractive drug target for treating obesity, diabetes, and eating disorders. However, the lack of structural information has hindered our understanding of the peptide recognition and regulatory mechanism of QRFPR, impeding drug design efforts. In this study, we determined the cryo-EM structure of the G q -coupled QRFPR bound to 26RFa. The structure reveals a unique assembly mode of the extracellular region of the receptor and the N-terminus of the peptide, and elucidates the recognition mechanism of the C-terminal heptapeptide of 26RFa by the transmembrane binding pocket of QRFPR. The study also clarifies the similarities and distinctions in the binding pattern of the RF-amide moiety in five RF-amide peptides and the RY-amide segment in neuropeptide Y. These findings deepen our understanding of the RF-amide peptide recognition, aiding in the rational design of drugs targeting QRFPR and other RF-amide peptide receptors.
- Lingang Laboratory, Shanghai, China.
Organizational Affiliation: