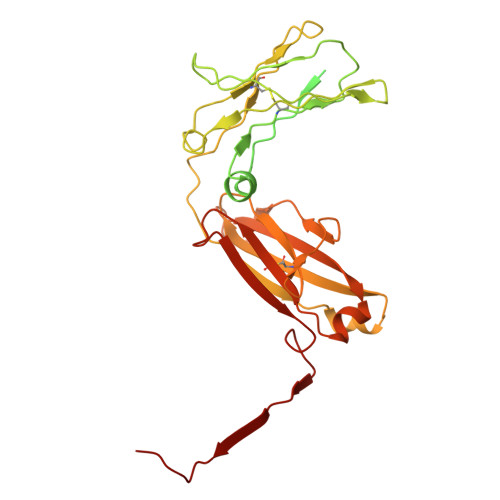
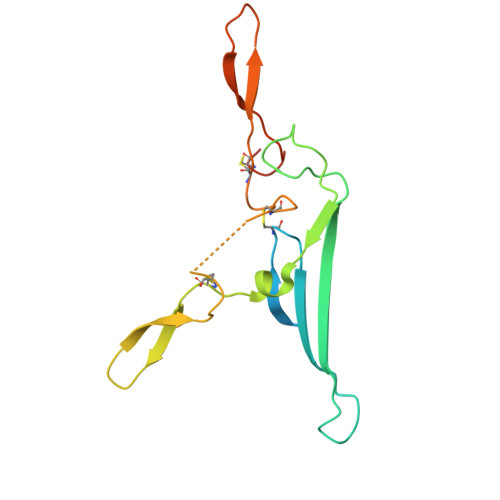
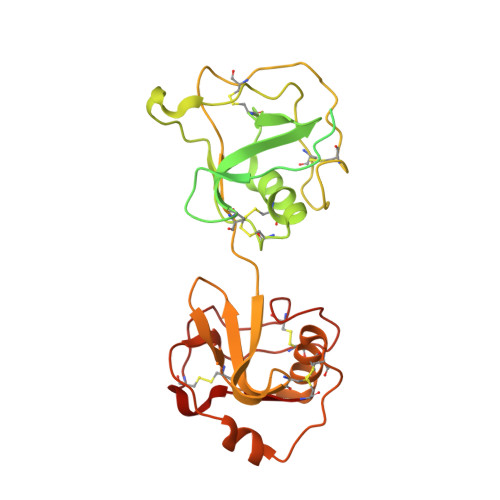
CD5L associates with IgM via the J chain.
Wang, Y., Su, C., Ji, C., Xiao, J.(2024) Nat Commun 15: 8397-8397
- PubMed: 39333069
- DOI: https://doi.org/10.1038/s41467-024-52175-y
- Primary Citation of Related Structures:
8WYR, 8WYS - PubMed Abstract:
CD5 antigen-like (CD5L), also known as Spα or AIM (Apoptosis inhibitor of macrophage), emerges as an integral component of serum immunoglobulin M (IgM). However, the molecular mechanism underlying the interaction between IgM and CD5L has remained elusive. In this study, we present a cryo-electron microscopy structure of the human IgM pentamer core in complex with CD5L. Our findings reveal that CD5L binds to the joining chain (J chain) in a Ca 2+ -dependent manner and further links to IgM via a disulfide bond. We further corroborate recently published data that CD5L reduces IgM binding to the mucosal transport receptor pIgR, but does not impact the binding of the IgM-specific receptor FcμR. Additionally, CD5L does not interfere with IgM-mediated complement activation. These results offer a more comprehensive understanding of IgM and shed light on the function of the J chain in the immune system.
- State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, Beijing, P.R. China.
Organizational Affiliation: