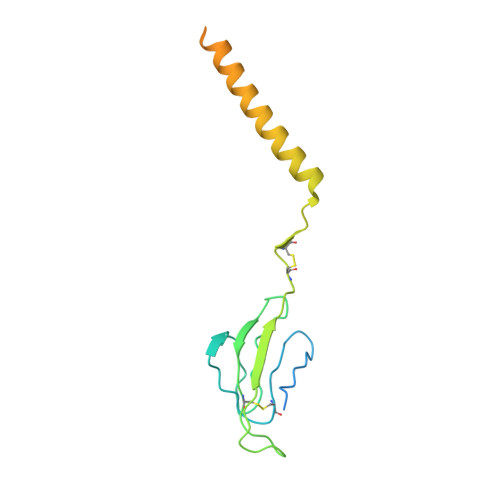
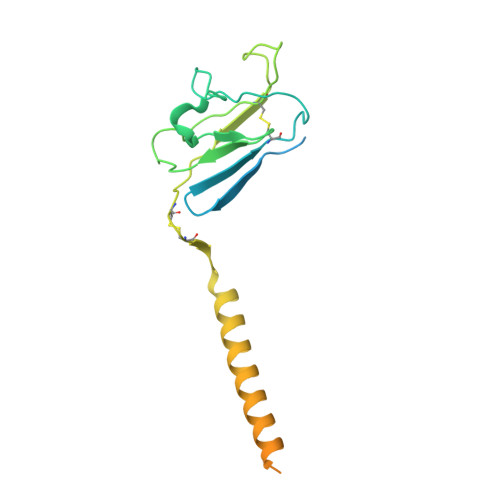
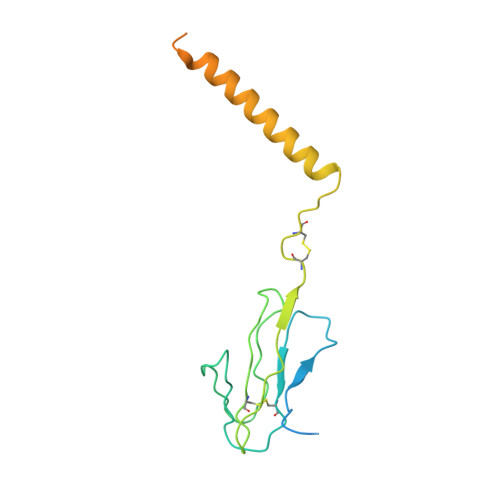
Structures of human gamma delta T cell receptor-CD3 complex.
Xin, W., Huang, B., Chi, X., Liu, Y., Xu, M., Zhang, Y., Li, X., Su, Q., Zhou, Q.(2024) Nature 630: 222-229
- PubMed: 38657677
- DOI: https://doi.org/10.1038/s41586-024-07439-4
- Primary Citation of Related Structures:
8JBV, 8JC0, 8JCB, 8WXE, 8WY0, 8WYI, 8YC0 - PubMed Abstract:
Gamma delta (γδ) T cells, a unique T cell subgroup, are crucial in various immune responses and immunopathology 1-3 . The γδ T cell receptor (TCR), generated by γδ T cells, recognizes a diverse range of antigens independently of the major histocompatibility complex 2 . The γδ TCR associates with CD3 subunits, initiating T cell activation and holding great potential in immunotherapy 4 . Here, we report the structures of two prototypical human Vγ9Vδ2 and Vγ5Vδ1 TCR-CD3 complexes 5,6 , unveiling two distinct assembly mechanisms that depend on Vγ usage. The Vγ9Vδ2 TCR-CD3 complex is monomeric, with considerable conformational flexibility in the TCRγ/TCRδ extracellular domain (ECD) and connecting peptides (CPs). The length of CPs regulates the ligand association and T cell activation. Additionally, a cholesterol-like molecule wedges into the transmembrane region, exerting an inhibitory role in TCR signaling. The Vγ5Vδ1 TCR-CD3 complex displays a dimeric architecture, where two protomers nestle back-to-back via their Vγ5 domains of TCR ECDs. Our biochemical and biophysical assays further corroborate the dimeric structure. Importantly, the dimeric form of the Vγ5Vδ1 TCR is essential for T cell activation. These findings reveal organizing principles of the γδ TCR-CD3 complex, providing insights into the γδ TCR unique properties and facilitating immunotherapeutic interventions.
- Research Center for Industries of the Future, Center for Infectious Disease Research, Zhejiang Key Laboratory of Structural Biology, School of Life Sciences, Westlake University; Institute of Biology, Westlake Institute for Advanced Study, Hangzhou, Zhejiang Province, China.
Organizational Affiliation: