Structure of ADP-Form AsfvPrimPol Hexamer
Shi, T.H., Guo, Y.Y., Yan, R.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative primase C962R | 972 | African swine fever virus | Mutation(s): 0 Gene Names: C962R CDS | 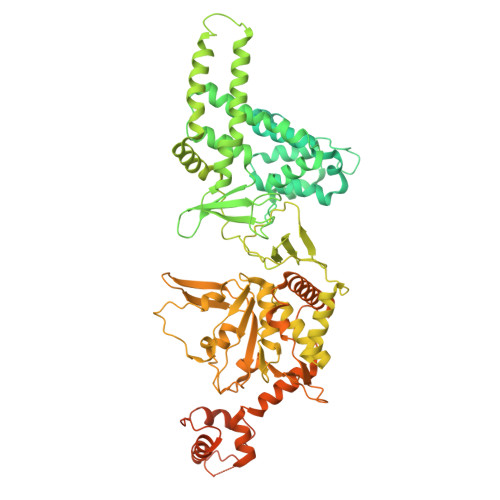 | |
UniProt | |||||
Find proteins for A0A2X0TKI6 (African swine fever virus) Explore A0A2X0TKI6 Go to UniProtKB: A0A2X0TKI6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2X0TKI6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (26-MER) | A [auth H] | 26 | Homo sapiens | 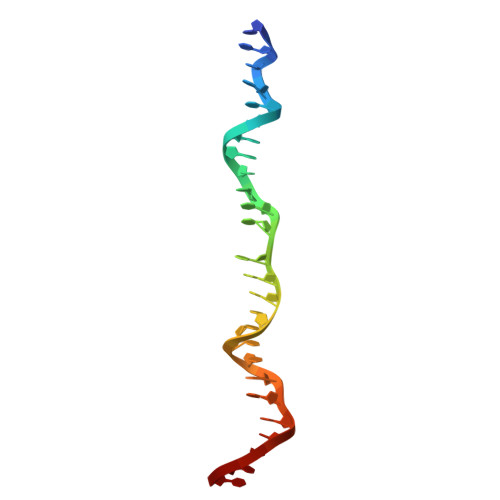 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*AP*AP*AP*A)-3') | B [auth I] | 4 | Homo sapiens | 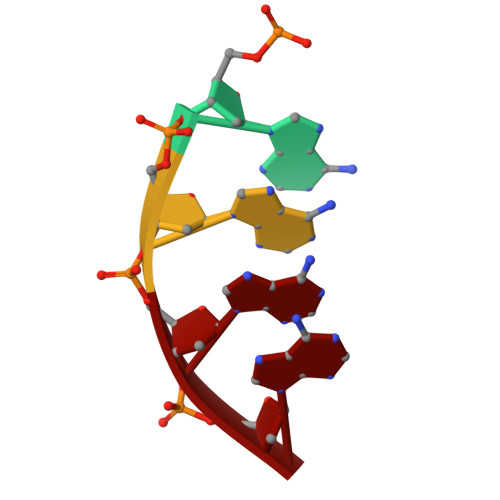 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ANP Query on ANP | I [auth A] K [auth B] M [auth D] O [auth E] Q [auth F] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| MG Query on MG | J [auth A] L [auth B] N [auth D] P [auth E] R [auth F] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |