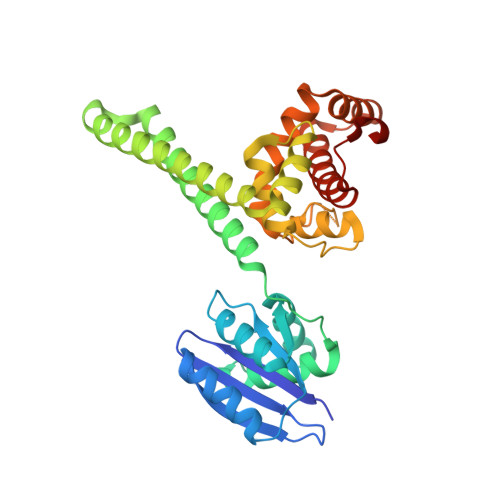
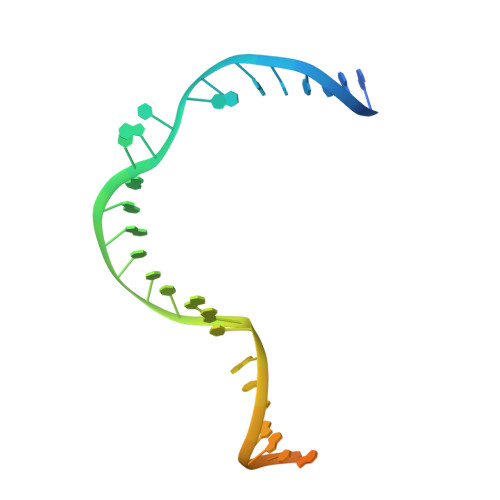
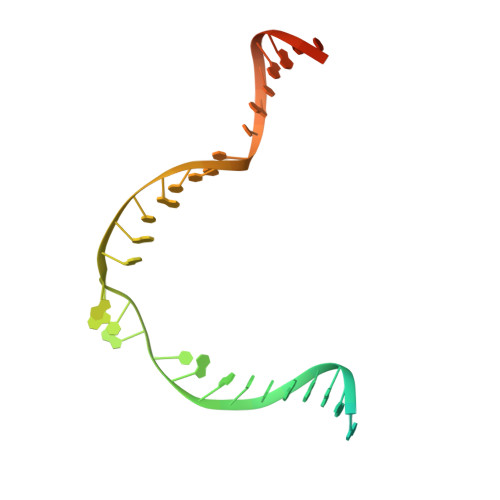
Structural mechanism of bridge RNA-guided recombination.
Hiraizumi, M., Perry, N.T., Durrant, M.G., Soma, T., Nagahata, N., Okazaki, S., Athukoralage, J.S., Isayama, Y., Pai, J.J., Pawluk, A., Konermann, S., Yamashita, K., Hsu, P.D., Nishimasu, H.(2024) Nature 630: 994-1002
- PubMed: 38926616
- DOI: https://doi.org/10.1038/s41586-024-07570-2
- Primary Citation of Related Structures:
8WT6, 8WT7, 8WT8, 8WT9 - PubMed Abstract:
Insertion sequence (IS) elements are the simplest autonomous transposable elements found in prokaryotic genomes 1 . We recently discovered that IS110 family elements encode a recombinase and a non-coding bridge RNA (bRNA) that confers modular specificity for target DNA and donor DNA through two programmable loops 2 . Here we report the cryo-electron microscopy structures of the IS110 recombinase in complex with its bRNA, target DNA and donor DNA in three different stages of the recombination reaction cycle. The IS110 synaptic complex comprises two recombinase dimers, one of which houses the target-binding loop of the bRNA and binds to target DNA, whereas the other coordinates the bRNA donor-binding loop and donor DNA. We uncovered the formation of a composite RuvC-Tnp active site that spans the two dimers, positioning the catalytic serine residues adjacent to the recombination sites in both target and donor DNA. A comparison of the three structures revealed that (1) the top strands of target and donor DNA are cleaved at the composite active sites to form covalent 5'-phosphoserine intermediates, (2) the cleaved DNA strands are exchanged and religated to create a Holliday junction intermediate, and (3) this intermediate is subsequently resolved by cleavage of the bottom strands. Overall, this study reveals the mechanism by which a bispecific RNA confers target and donor DNA specificity to IS110 recombinases for programmable DNA recombination.
- Department of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: