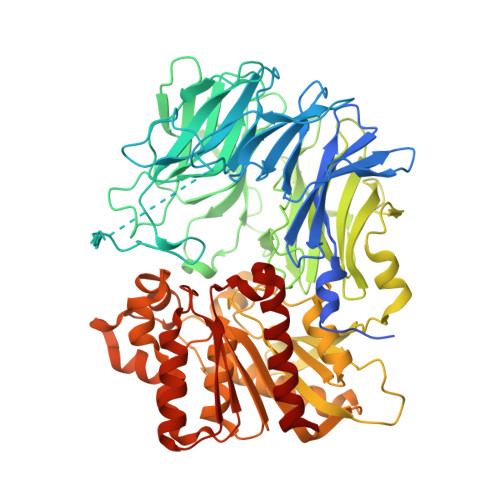
Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Chandravanshi, K., Singh, R., Bhange, G.N., Kumar, A., Yadav, P., Kumar, A., Makde, R.D.(2024) FEBS Lett 598: 684-701
- PubMed: 38426217
- DOI: https://doi.org/10.1002/1873-3468.14834
- Primary Citation of Related Structures:
8WT1 - PubMed Abstract:
Acylaminoacyl peptidases (AAPs) play a pivotal role in various pathological conditions and are recognized as potential therapeutic targets. AAPs exhibit a wide range of activities, such as acylated amino acid-dependent aminopeptidase, endopeptidase, and less studied carboxypeptidase activity. We have determined the crystal structure of an AAP from Geobacillus stearothermophilus (S9gs) at 2.0 Å resolution. Despite being annotated as an aminopeptidase in the NCBI database, our enzymatic characterization proved S9gs to be a carboxypeptidase. Solution-scattering studies showed that S9gs exists as a tetramer in solution, and crystal structure analysis revealed adaptations responsible for the carboxypeptidase activity of S9gs. The findings present a hypothesis for substrate selection, substrate entry, and product exit from the active site, enriching our understanding of this rare carboxypeptidase.
Organizational Affiliation:
Department of Bioscience and Biomedical Engineering, Indian Institute of Technology Indore, India.