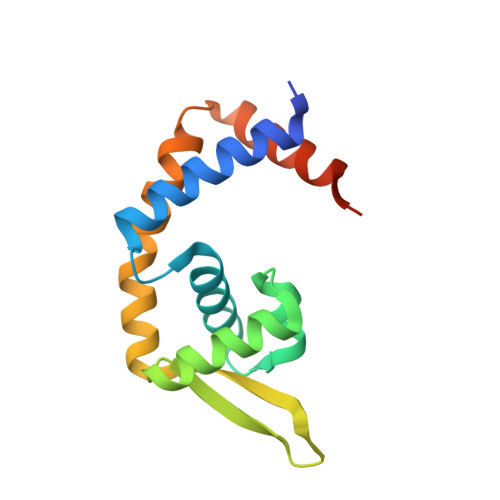
Pre-binding structure of HosA transcriptional regulator from enteropathogenic Escherichia coli O127:H6 (strain E2348/69) in presence of sub optimal concentration of 4-hydroxy benzoic acid
Arpita, G., Rukmini, R., Mallesham, K., Samee, U.(2024) bioRxiv