Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
Gimenez, L.E., Martin, C., Yu, J., Hollanders, C., Hernandez, C.C., Wu, Y., Yao, D., Han, G.W., Dahir, N.S., Wu, L., Van der Poorten, O., Lamouroux, A., Mannes, M., Zhao, S., Tourwe, D., Stevens, R.C., Cone, R.D., Ballet, S.(2024) J Med Chem 67: 2690-2711
- PubMed: 38345933
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01822
- Primary Citation of Related Structures:
8WKY, 8WKZ - PubMed Abstract:
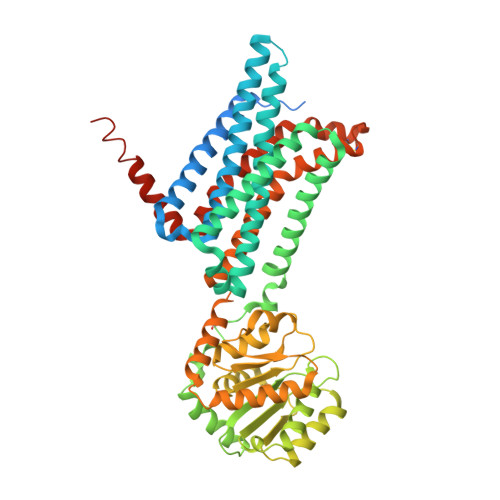
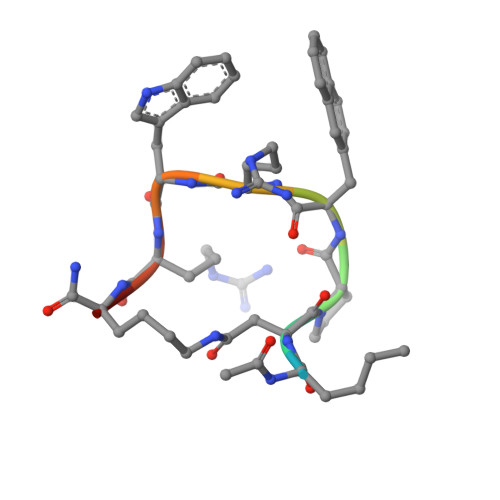
Melanocortin 4 receptor (MC4-R) antagonists are actively sought for treating cancer cachexia. We determined the structures of complexes with PG-934 and SBL-MC-31 . These peptides differ from SHU9119 by substituting His 6 with Pro 6 and inserting Gly 10 or Arg 10 . The structures revealed two subpockets at the TM7-TM1-TM2 domains, separated by N285 7.36 . Two peptide series based on the complexed peptides led to an antagonist activity and selectivity SAR study. Most ligands retained the SHU9119 potency, but several SBL-MC-31 -derived peptides significantly enhanced MC4-R selectivity over MC1-R by 60- to 132-fold. We also investigated MC4-R coupling to the K + channel, Kir7.1. Some peptides activated the channel, whereas others induced channel closure independently of G protein coupling. In cell culture studies, channel activation correlated with increased feeding, while a peptide with Kir7.1 inhibitory activity reduced eating. These results highlight the potential for targeting the MC4-R:Kir7.1 complex for treating positive and restrictive eating disorders.
- Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, United States.
Organizational Affiliation: