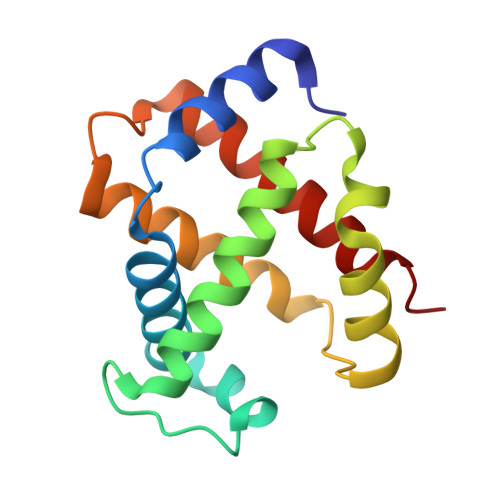
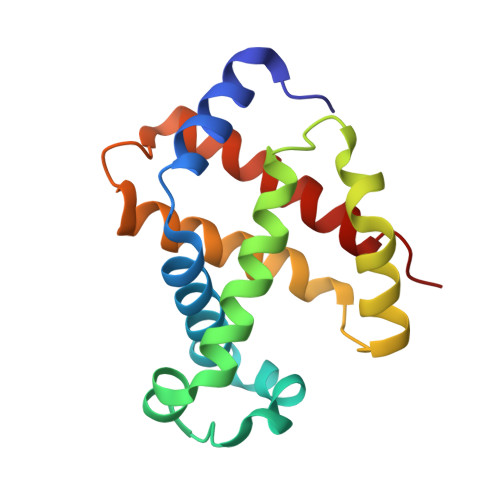
The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Takahashi, K., Lee, Y., Fago, A., Bautista, N.M., Storz, J.F., Kawamoto, A., Kurisu, G., Nishizawa, T., Tame, J.R.H.(2024) Nat Commun 15: 6505-6505
- PubMed: 39090102
- DOI: https://doi.org/10.1038/s41467-024-49947-x
- Primary Citation of Related Structures:
8WIX, 8WIY, 8WIZ, 8WJ0, 8WJ1, 8WJ2 - PubMed Abstract:
The principal effect controlling the oxygen affinity of vertebrate haemoglobins (Hbs) is the allosteric switch between R and T forms with relatively high and low oxygen affinity respectively. Uniquely among jawed vertebrates, crocodilians possess Hb that shows a profound drop in oxygen affinity in the presence of bicarbonate ions. This allows them to stay underwater for extended periods by consuming almost all the oxygen present in the blood-stream, as metabolism releases carbon dioxide, whose conversion to bicarbonate and hydrogen ions is catalysed by carbonic anhydrase. Despite the apparent universal utility of bicarbonate as an allosteric regulator of Hb, this property evolved only in crocodilians. We report here the molecular structures of both human and a crocodilian Hb in the deoxy and liganded states, solved by cryo-electron microscopy. We reveal the precise interactions between two bicarbonate ions and the crocodilian protein at symmetry-related sites found only in the T state. No other known effector of vertebrate Hbs binds anywhere near these sites.
- Graduate School of Medical Life Science, Yokohama City University, Suehiro 1-7-29, Yokohama, 230-0045, Japan.
Organizational Affiliation: