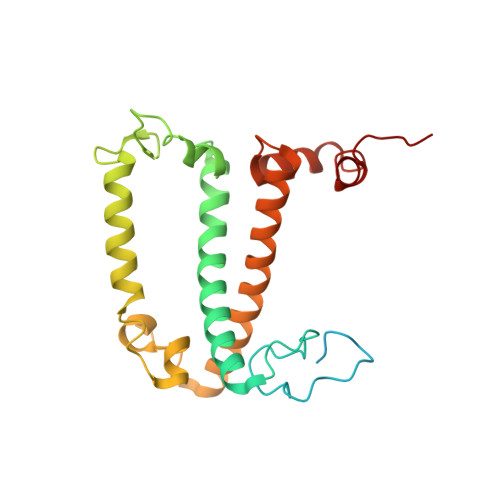
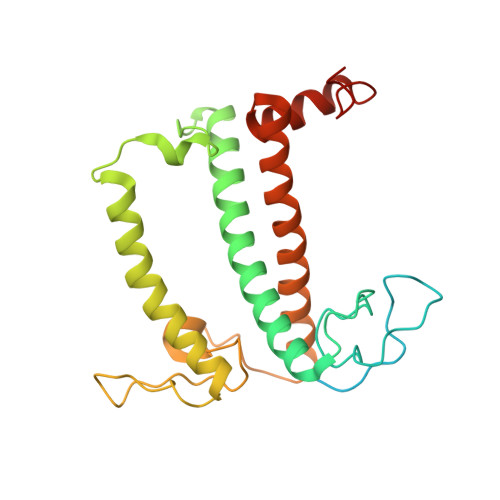
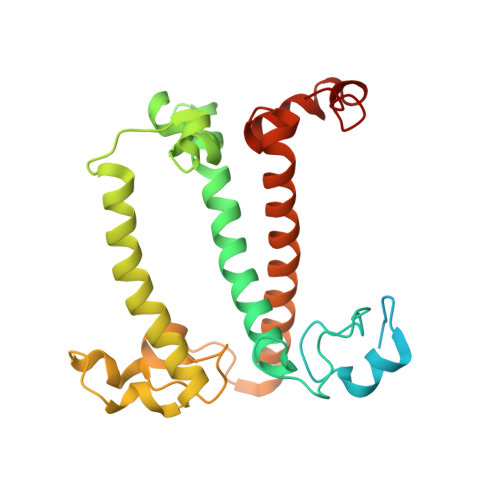
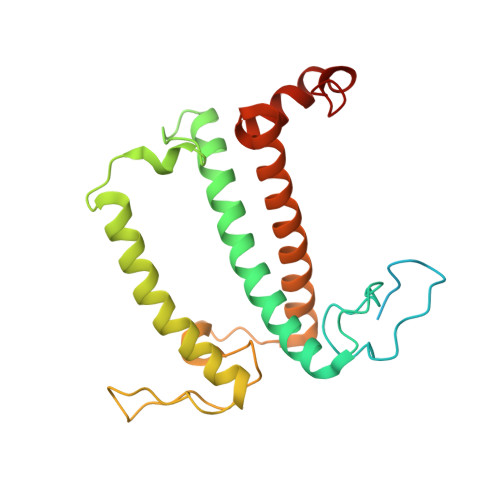
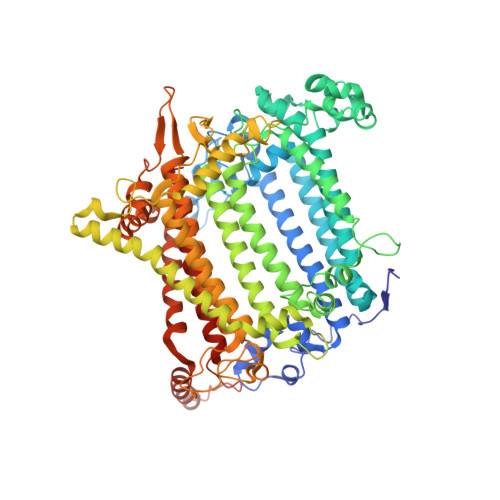
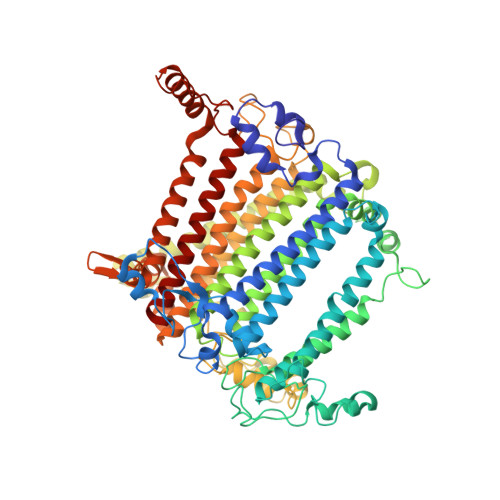
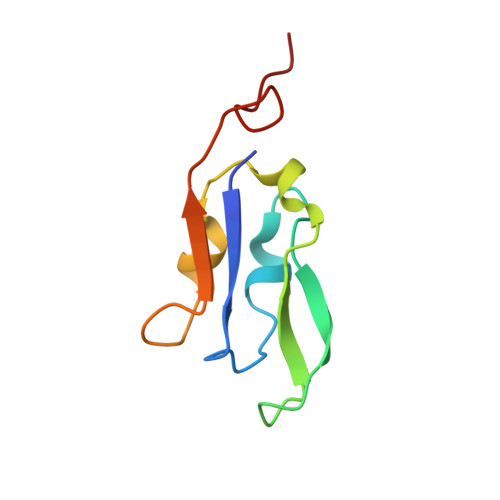
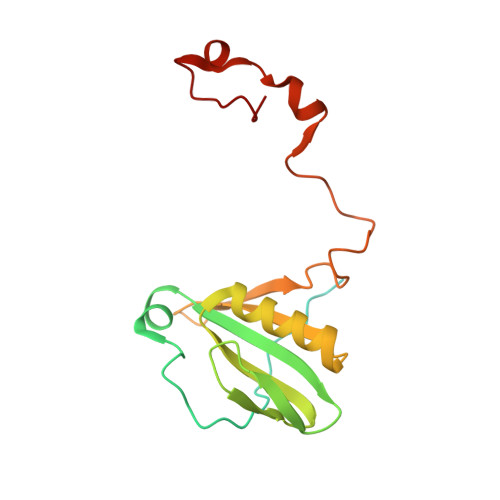
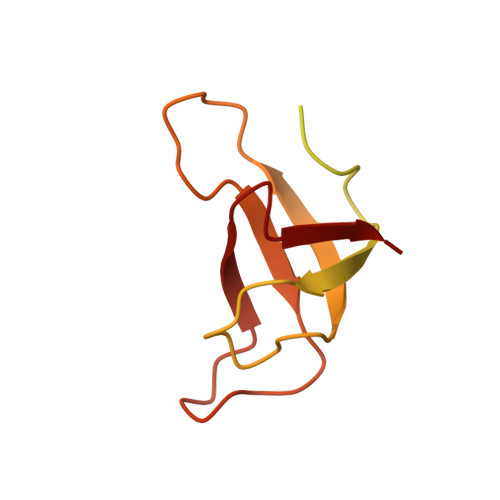
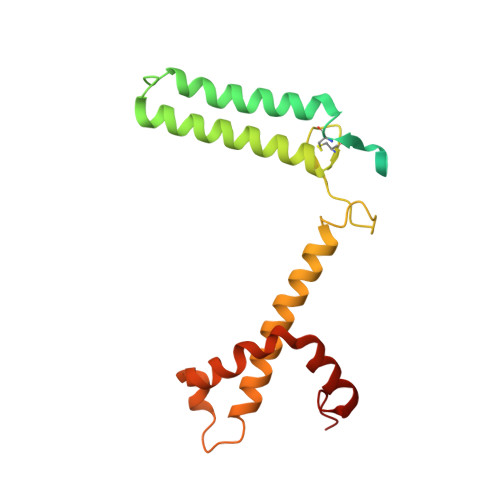
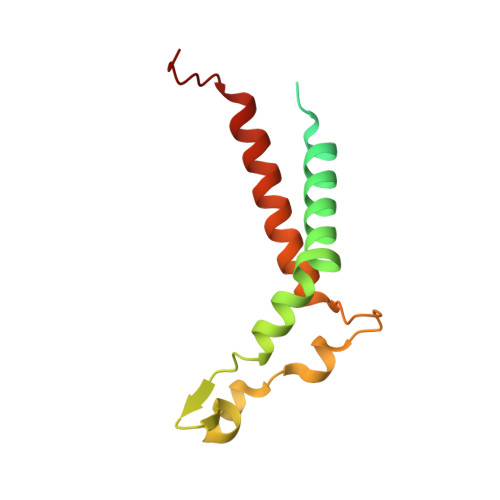
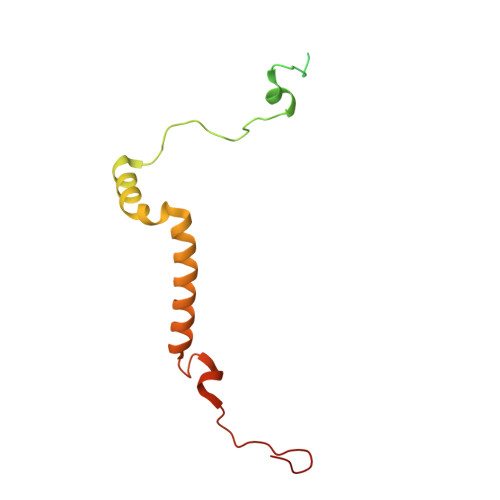
Structure of the red-shifted Fittonia albivenis photosystem I.
Li, X., Huang, G., Zhu, L., Hao, C., Sui, S.F., Qin, X.(2024) Nat Commun 15: 6325-6325
- PubMed: 39060282
- DOI: https://doi.org/10.1038/s41467-024-50655-9
- Primary Citation of Related Structures:
8WGH - PubMed Abstract:
Photosystem I (PSI) from Fittonia albivenis, an Acanthaceae ornamental plant, is notable among green plants for its red-shifted emission spectrum. Here, we solved the structure of a PSI-light harvesting complex I (LHCI) supercomplex from F. albivenis at 2.46-Å resolution using cryo-electron microscopy. The supercomplex contains a core complex of 14 subunits and an LHCI belt with four antenna subunits (Lhca1-4) similar to previously reported angiosperm PSI-LHCI structures; however, Lhca3 differs in three regions surrounding a dimer of low-energy chlorophylls (Chls) termed red Chls, which absorb far-red beyond visible light. The unique amino acid sequences within these regions are exclusively shared by plants with strongly red-shifted fluorescence emission, suggesting candidate structural elements for regulating the energy state of red Chls. These results provide a structural basis for unraveling the mechanisms of light harvest and transfer in PSI-LHCI of under canopy plants and for designing Lhc to harness longer-wavelength light in the far-red spectral range.
- School of Chemistry and Chemical Engineering, School of Biological Science and Technology, University of Jinan, Jinan, China.
Organizational Affiliation: