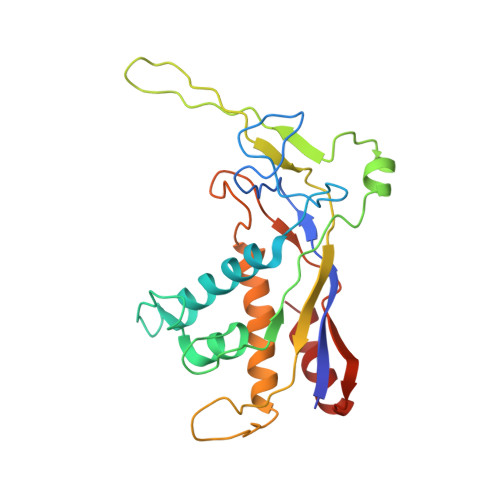
Type-III-A structure of mycobacteria CRISPR-Csm complexes involving atypical crRNAs.
Zhang, H., Shi, M., Ma, X., Liu, M., Wang, N., Lu, Q., Li, Z., Zhao, Y., Zhao, H., Chen, H., Zhang, H., Jiang, T., Ouyang, S., Huo, Y., Bi, L.(2024) Int J Biol Macromol 260: 129331-129331
- PubMed: 38218299
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.129331
- Primary Citation of Related Structures:
8WFX, 8X5D - PubMed Abstract:
Tuberculosis (TB), a leading cause of mortality globally, is a chronic infectious disease caused by Mycobacterium tuberculosis that primarily infiltrates the lung. The mature crRNAs in M. tuberculosis transcribed from the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) locus exhibit an atypical structure featured with 5' and 3' repeat tags at both ends of the intact crRNA, in contrast to typical Type-III-A crRNAs that possess 5' repeat tags and partial crRNA sequences. However, this structural peculiarity particularly concerning the specific binding characteristics of the 3' repeat end within the mature crRNA within the Csm complex, has not been comprehensively elucidated. Here, our Mycobacteria CRISPR-Csm complexes structure represents the largest Csm complex reported to date. It incorporates an atypical Type-III-A CRISPR RNA (crRNA) (46 nt) with 5' 8-nt and 3' 4-nt repeat sequences in the stoichiometry of Mycobacteria Csm1 1 2 5 3 6 4 1 5 1. The PAM-independent single-stranded RNAs (ssRNAs) are the most suitable substrate for the Csm complex. The 3'-repeat end trimming of mature crRNA was not necessary for its cleavage activity in Type-III-A Csm complex. Our work broadens our understanding of the Type-III-A Csm complex and identifies another mature crRNA processing mechanism in the Type-III-A CRISPR-Cas system based on structural biology.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; Beijing Center for Disease Prevention and Control, Beijing 100013, China.
Organizational Affiliation: