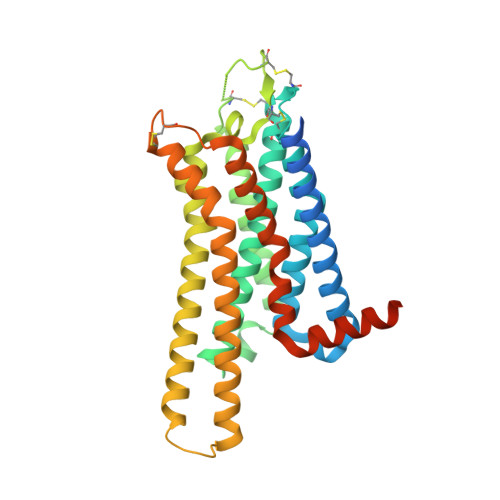
Crystal structure reveals the binding mode and selectivity of a photoswitchable ligand for the adenosine A 2A receptor.
Araya, T., Matsuba, Y., Suzuki, H., Doura, T., Nuemket, N., Nango, E., Yamamoto, M., Im, D., Asada, H., Kiyonaka, S., Iwata, S.(2023) Biochem Biophys Res Commun 695: 149393-149393
- PubMed: 38171234
- DOI: https://doi.org/10.1016/j.bbrc.2023.149393
- Primary Citation of Related Structures:
8WDT - PubMed Abstract:
Rational synthetic expansion of photoresponsive ligands is important for photopharmacological studies. Adenosine A 2A receptor (A 2A R) is stimulated by adenosine and related in Parkinson's disease and other diseases. Here, we report the crystal structure of the A 2A R in complex with the novel photoresponsive ligand photoNECA (blue) at 3.34 Å resolution. PhotoNECA (blue) was designed for this structural study and the cell-based assay showed a photoresponsive and receptor selective characteristics of photoNECA (blue) for A 2A R. The crystal structure explains the binding mode, photoresponsive mechanism and receptor selectivity of photoNECA (blue). Our study would promote not only the rational design of photoresponsive ligands but also dynamic structural studies of A 2A R.
- Department of Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto, 606-8501, Japan.
Organizational Affiliation: