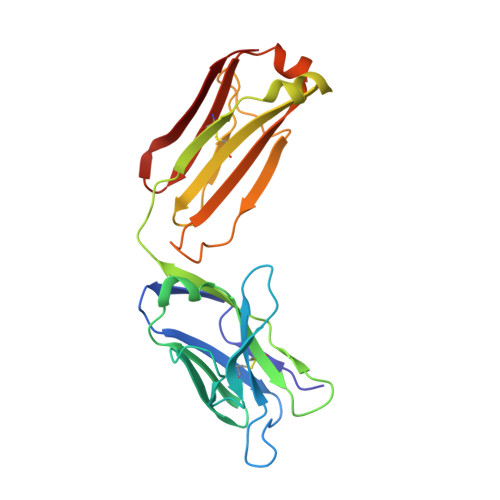
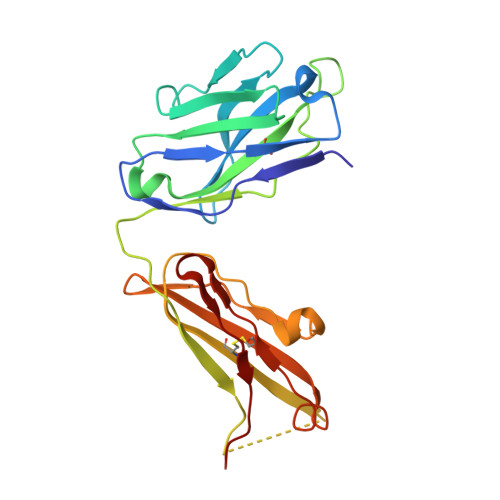
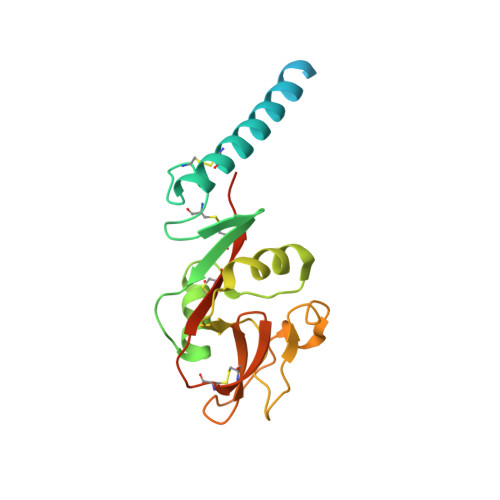
Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Mori, S., Nagae, M., Yamasaki, S.(2024) Int Immunol 36: 279-290
- PubMed: 38386511
- DOI: https://doi.org/10.1093/intimm/dxae006
- Primary Citation of Related Structures:
8W8T, 8W9H, 8W9J - PubMed Abstract:
C-type lectin receptors (CLRs) are a family of pattern recognition receptors, which detect a broad spectrum of ligands via small carbohydrate-recognition domains (CRDs). CLEC12A is an inhibitory CLR that recognizes crystalline structures such as monosodium urate crystals. CLEC12A also recognizes mycolic acid, a major component of mycobacterial cell walls, and suppresses host immune responses. Although CLEC12A could be a therapeutic target for mycobacterial infection, structural information on CLEC12A was not available. We report here the crystal structures of human CLEC12A (hCLEC12A) in ligand-free form and in complex with 50C1, its inhibitory antibody. 50C1 recognizes human-specific residues on the top face of hCLEC12A CRD. A comprehensive alanine scan demonstrated that the ligand-binding sites of mycolic acid and monosodium urate crystals may overlap with each other, suggesting that CLEC12A utilizes a common interface to recognize different types of ligands. Our results provide atomic insights into the blocking and ligand-recognition mechanisms of CLEC12A and leads to the design of CLR-specific inhibitors.
- Department of Molecular Immunology, Research Institute for Microbial Diseases, Osaka University, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: