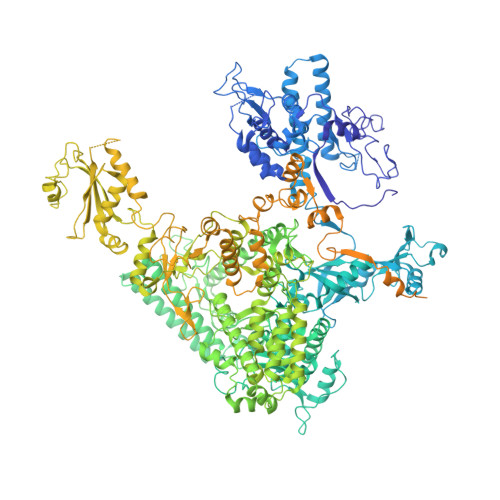
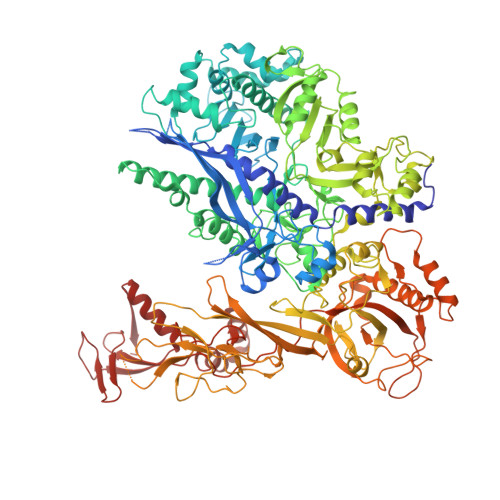
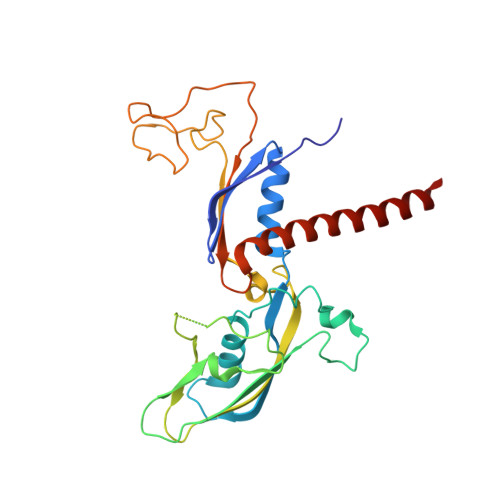
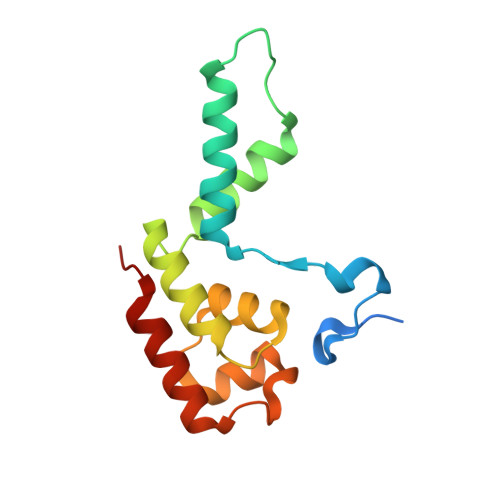
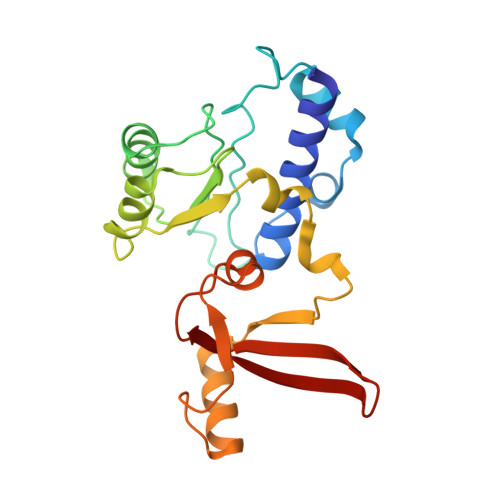
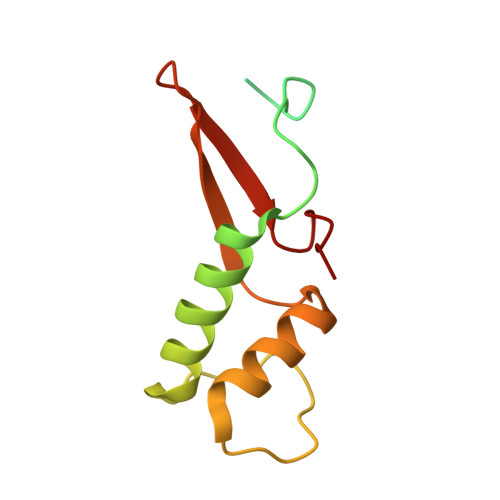
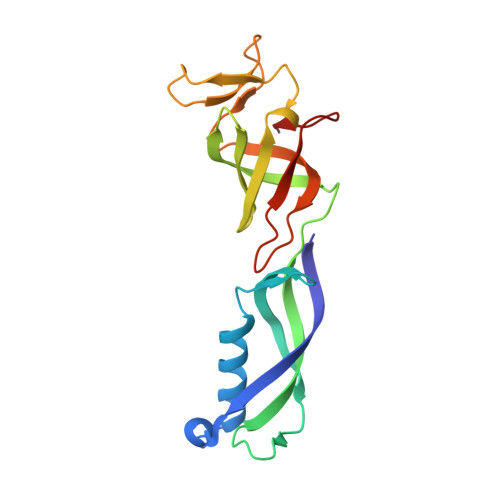
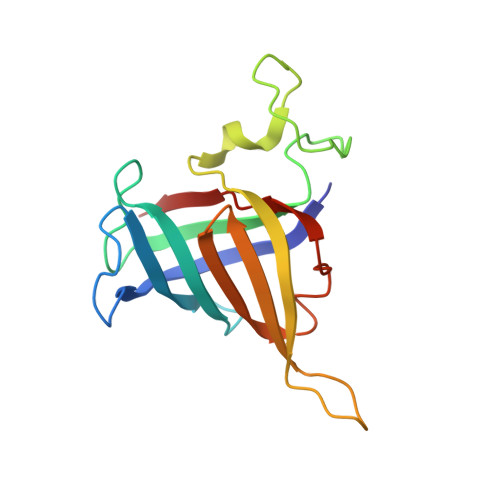
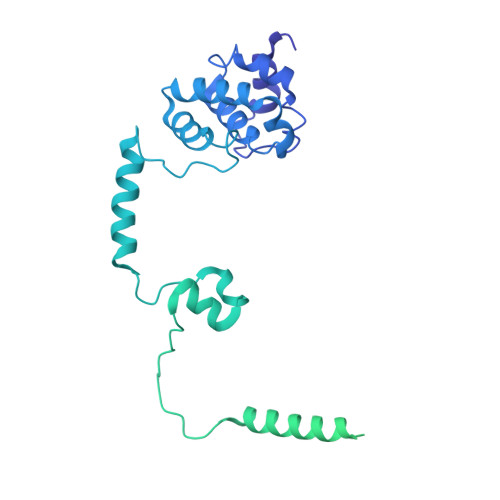
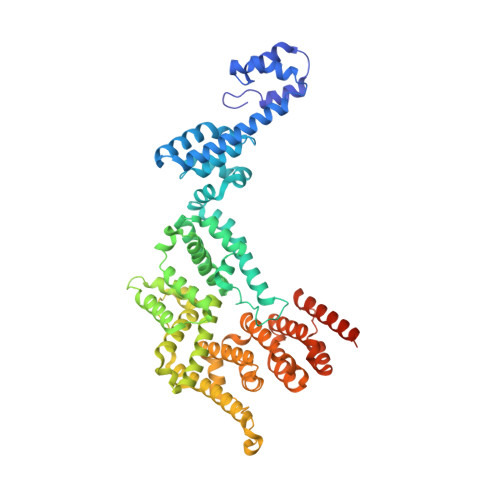
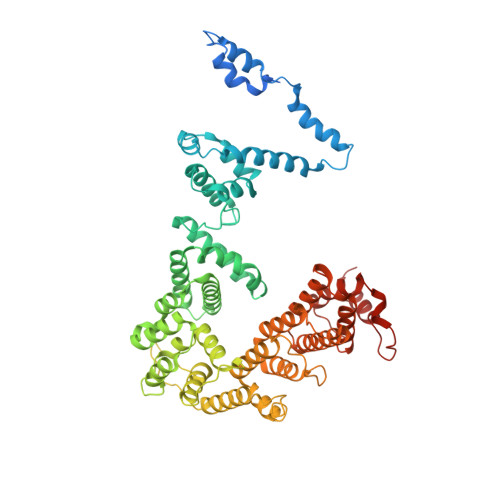
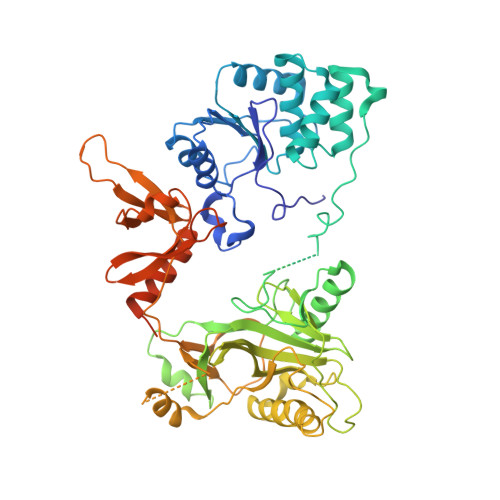
Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Li, Y., Wang, Q., Xu, Y., Li, Z.(2024) Nat Commun 15: 4622-4622
- PubMed: 38816438
- DOI: https://doi.org/10.1038/s41467-024-48963-1
- Primary Citation of Related Structures:
8W8E, 8W8F - PubMed Abstract:
The 5'-end capping of nascent pre-mRNA represents the initial step in RNA processing, with evidence demonstrating that guanosine addition and 2'-O-ribose methylation occur in tandem with early steps of transcription by RNA polymerase II, especially at the pausing stage. Here, we determine the cryo-EM structures of the paused elongation complex in complex with RNGTT, as well as the paused elongation complex in complex with RNGTT and CMTR1. Our findings show the simultaneous presence of RNGTT and the NELF complex bound to RNA polymerase II. The NELF complex exhibits two conformations, one of which shows a notable rearrangement of NELF-A/D compared to that of the paused elongation complex. Moreover, CMTR1 aligns adjacent to RNGTT on the RNA polymerase II stalk. Our structures indicate that RNGTT and CMTR1 directly bind the paused elongation complex, illuminating the mechanism by which 5'-end capping of pre-mRNA during transcriptional pausing.
- Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, State Key Laboratory of Genetic Engineering and Shanghai Key Laboratory of Medical Epigenetics, Shanghai Medical College of Fudan University, Shanghai, 200032, China.
Organizational Affiliation: