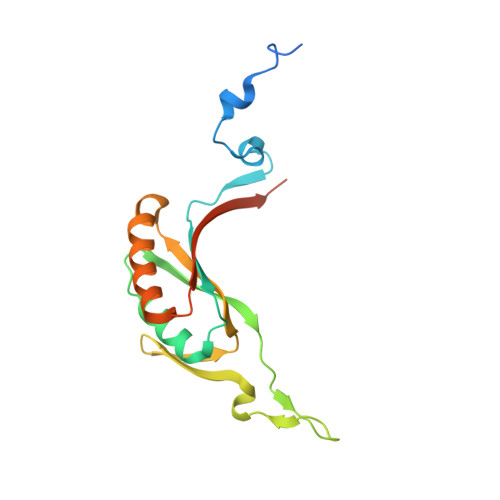
The crystal structure of methanogen McrD, a methyl-coenzyme M reductase-associated protein.
Sutherland-Smith, A.J., Carbone, V., Schofield, L.R., Cronin, B., Duin, E.C., Ronimus, R.S.(2024) FEBS Open Bio 14: 1222-1229
- PubMed: 38877345
- DOI: https://doi.org/10.1002/2211-5463.13848
- Primary Citation of Related Structures:
8W33 - PubMed Abstract:
Methyl-coenzyme M reductase (MCR) is a multi-subunit (α 2 β 2 γ 2 ) enzyme responsible for methane formation via its unique F 430 cofactor. The genes responsible for producing MCR (mcrA, mcrB and mcrG) are typically colocated with two other highly conserved genes mcrC and mcrD. We present here the high-resolution crystal structure for McrD from a human gut methanogen Methanomassiliicoccus luminyensis strain B10. The structure reveals that McrD comprises a ferredoxin-like domain assembled into an α + β barrel-like dimer with conformational flexibility exhibited by a functional loop. The description of the M. luminyensis McrD crystal structure contributes to our understanding of this key conserved methanogen protein typically responsible for promoting MCR activity and the production of methane, a greenhouse gas.
Organizational Affiliation:
School of Natural Sciences, Massey University, Palmerston North, New Zealand.