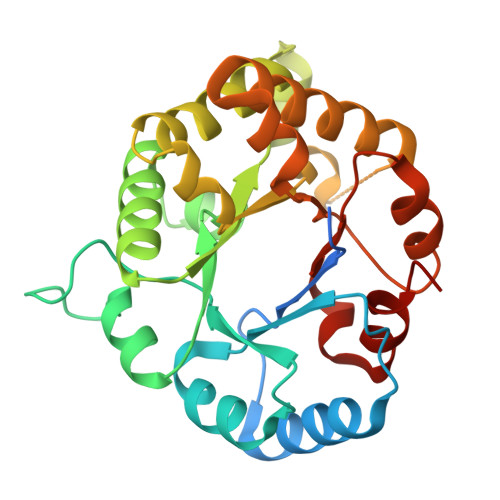
Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Perez-Nino, J.A., Guerra, Y., Diaz-Salazar, A.J., Costas, M., Rodriguez-Romero, A., Fernandez-Velasco, D.A.(2024) Protein Sci 33: e5134-e5134
- PubMed: 39145435
- DOI: https://doi.org/10.1002/pro.5134
- Primary Citation of Related Structures:
8V09, 8V0A, 8V2W, 8V2X, 8W05, 8W06, 8W08 - PubMed Abstract:
Function and structure are strongly coupled in obligated oligomers such as Triosephosphate isomerase (TIM). In animals and fungi, TIM monomers are inactive and unstable. Previously, we used ancestral sequence reconstruction to study TIM evolution and found that before these lineages diverged, the last opisthokonta common ancestor of TIM (LOCATIM) was an obligated oligomer that resembles those of extant TIMs. Notably, calorimetric evidence indicated that ancestral TIM monomers are more structured than extant ones. To further increase confidence about the function, structure, and stability of the LOCATIM, in this work, we applied two different inference methodologies and the worst plausible case scenario for both of them, to infer four sequences of this ancestor and test the robustness of their physicochemical properties. The extensive biophysical characterization of the four reconstructed sequences of LOCATIM showed very similar hydrodynamic and spectroscopic properties, as well as ligand-binding energetics and catalytic parameters. Their 3D structures were also conserved. Although differences were observed in melting temperature, all LOCATIMs showed reversible urea-induced unfolding transitions, and for those that reached equilibrium, high conformational stability was estimated (ΔG Tot = 40.6-46.2 kcal/mol). The stability of the inactive monomeric intermediates was also high (ΔG unf = 12.6-18.4 kcal/mol), resembling some protozoan TIMs rather than the unstable monomer observed in extant opisthokonts. A comparative analysis of the 3D structure of ancestral and extant TIMs shows a correlation between the higher stability of the ancestral monomers with the presence of several hydrogen bonds located in the "bottom" part of the barrel.
- Laboratorio de Fisicoquímica e Ingeniería de Proteínas, Departamento de Bioquímica, Facultad de Medicina, Universidad Nacional Autónoma de México, Ciudad de México, Mexico.
Organizational Affiliation: