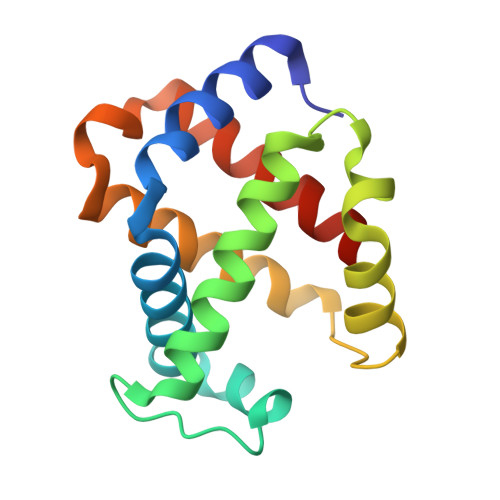
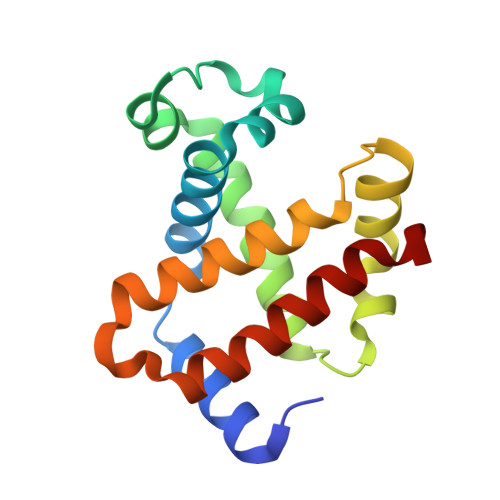
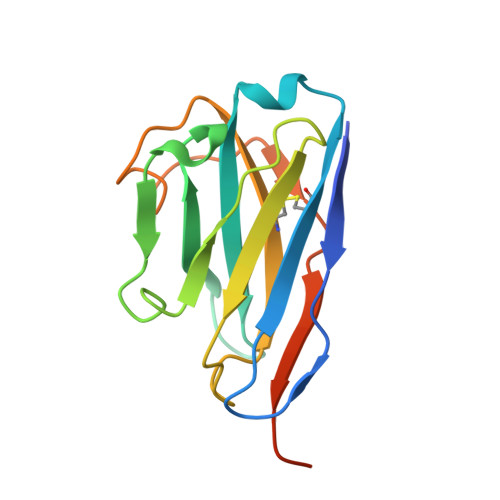
The structure of a haemoglobin-nanobody complex reveals human beta-subunit-specific interactions.
Fox, D.R., Samuels, I., Binks, S., Grinter, R.(2024) FEBS Lett 598: 2240-2248
- PubMed: 38880764
- DOI: https://doi.org/10.1002/1873-3468.14958
- Primary Citation of Related Structures:
8VYL - PubMed Abstract:
Haemoglobin (Hb) is a vital oxygen carrier in vertebrates. Low blood Hb levels may indicate anaemia or genetic disorders, while its presence in the lower digestive system suggests colon cancer. Detecting and quantifying human Hb is essential for medical diagnostics. A nanobody-based sandwich-ELISA test was recently developed utilising llama-derived nanobodies NbE11 and NbB9. These nanobodies specifically bind to human Hb without cross-reacting with Hb from other vertebrates. Here, we determine the crystal structure of NbE11 in complex with human Hb. NbE11 binds Hb with high affinity, predominantly binding the β-Hb subunit. Structural differences between human Hb and other vertebrates at the NbE11 binding interface likely explain the assay's lack of cross-reactivity, providing insights for developing Hb binding diagnostics.
Organizational Affiliation:
Department of Microbiology, Biomedicine Discovery Institute, Monash University, Clayton, Australia.