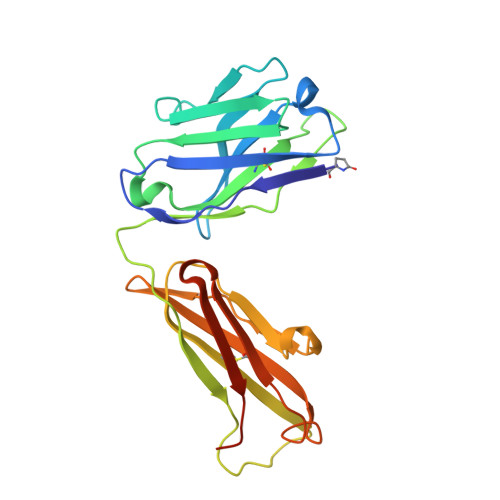
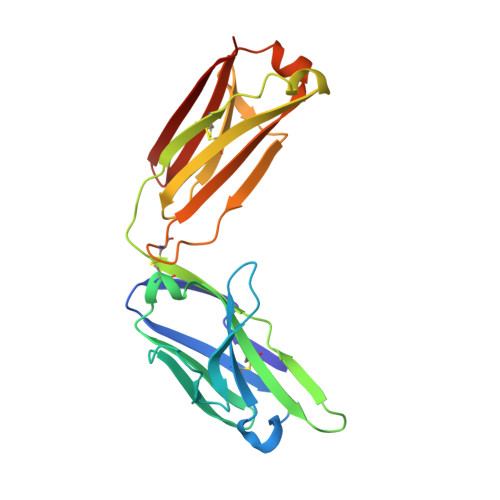
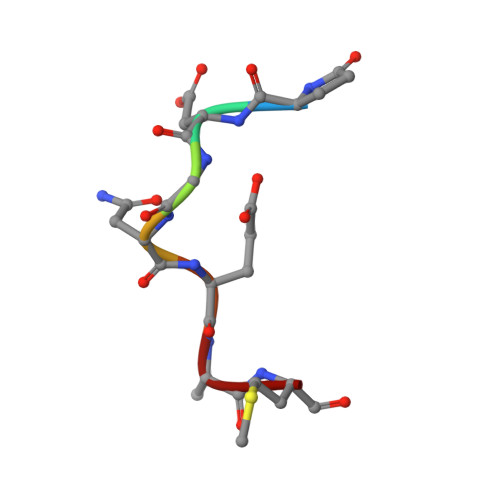
Engineering a tumor-selective prodrug T-cell engager bispecific antibody for safer immunotherapy.
McCue, A.C., Demarest, S.J., Froning, K.J., Hickey, M.J., Antonysamy, S., Kuhlman, B.(2024) MAbs 16: 2373325-2373325
- PubMed: 38962811
- DOI: https://doi.org/10.1080/19420862.2024.2373325
- Primary Citation of Related Structures:
8VY4 - PubMed Abstract:
T-cell engaging (TCE) bispecific antibodies are potent drugs that trigger the immune system to eliminate cancer cells, but administration can be accompanied by toxic side effects that limit dosing. TCEs function by binding to cell surface receptors on T cells, frequently CD3, with one arm of the bispecific antibody while the other arm binds to cell surface antigens on cancer cells. On-target, off-tumor toxicity can arise when the target antigen is also present on healthy cells. The toxicity of TCEs may be ameliorated through the use of pro-drug forms of the TCE, which are not fully functional until recruited to the tumor microenvironment. This can be accomplished by masking the anti-CD3 arm of the TCE with an autoinhibitory motif that is released by tumor-enriched proteases. Here, we solve the crystal structure of the antigen-binding fragment of a novel anti-CD3 antibody, E10, in complex with its epitope from CD3 and use this information to engineer a masked form of the antibody that can activate by the tumor-enriched protease matrix metalloproteinase 2 (MMP-2). We demonstrate with binding experiments and in vitro T-cell activation and killing assays that our designed prodrug TCE is capable of tumor-selective T-cell activity that is dependent upon MMP-2. Furthermore, we demonstrate that a similar masking strategy can be used to create a pro-drug form of the frequently used anti-CD3 antibody SP34. This study showcases an approach to developing immune-modulating therapeutics that prioritizes safety and has the potential to advance cancer immunotherapy treatment strategies.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC, USA.