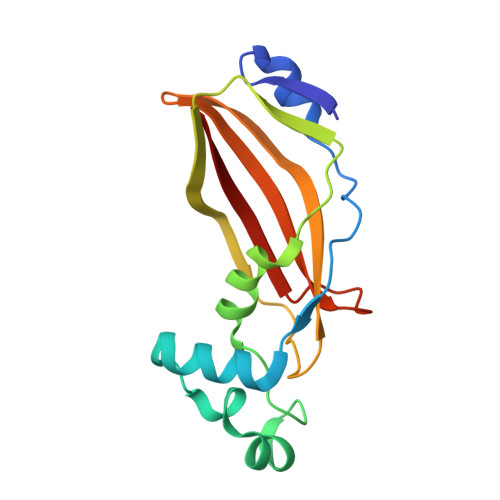
Shy is a proteobacterial steroid hydratase which catalyzes steroid side chain degradation without requiring a catalytically inert partner domain.
Schroeter, K.L., Rolfe, N., Forrester, T.J.B., Kimber, M.S., Seah, S.Y.K.(2024) J Biological Chem 300: 107509-107509
- PubMed: 38944126
- DOI: https://doi.org/10.1016/j.jbc.2024.107509
- Primary Citation of Related Structures:
8VWR - PubMed Abstract:
Shy (side chain hydratase) and Sal (side chain aldolase), are involved in successive reactions in the pathway of bile acid side chain catabolism in Proteobacteria. Untagged Shy co-purified with His-tagged Sal indicating that the two enzymes form a complex. Shy contains a MaoC and a DUF35 domain. When co-expressed with Sal, the DUF35 domain but not the MaoC domain of Shy was observed to co-purify with Sal, indicating Sal interacts with Shy through its DUF35 domain. The MaoC domain of Shy (Shy MaoC ) remained catalytically viable and could hydrate cholyl-enoyl-CoA with similar catalytic efficiency as in the Shy-Sal complex. Sal expressed with the DUF35 domain of Shy (Sal-Shy DUF35 ) was similarly competent for the retroaldol cleavage of cholyl-3-OH-CoA. Shy MaoC showed a preference for C 5 side chain bile acid substrates, exhibiting low activity towards C 3 side chain substrates. The Shy MaoC structure was determined by X-ray crystallography, showing a hot dog fold with a short central helix surrounded by a twisted anti-parallel β-sheet. Modeling and mutagenesis studies suggest that the bile acid substrate occupies the large open cleft formed by the truncated central helix and repositioning of the active site housing. Shy MaoC therefore contains two substrate binding sites per homodimer, making it distinct from previously characterized MaoC steroid hydratases that are (pseudo)-heterodimers with one substrate binding site per dimer. The characterization of Shy provides insight into how MaoC family hydratases have adapted to accommodate large polycyclic substrates that can facilitate future engineering of these enzymes to produce novel steroid pharmaceuticals.
- Department of Molecular and Cellular Biology, University of Guelph, Canada.
Organizational Affiliation: