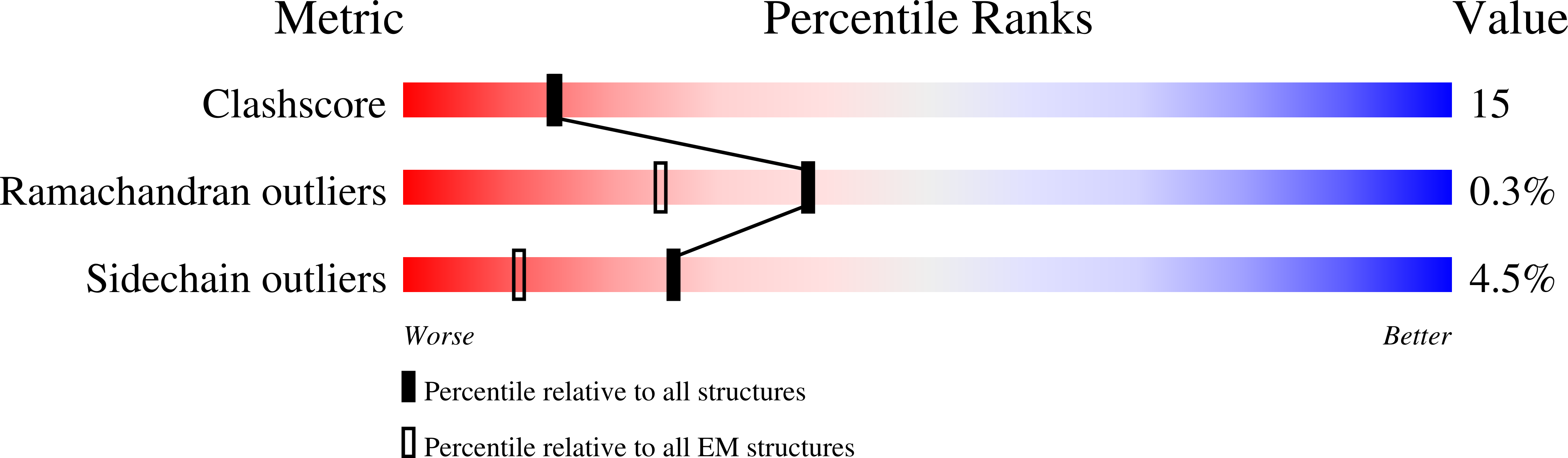Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Olia, A.S., Prabhakaran, M., Harris, D.R., Cheung, C.S., Gillespie, R.A., Gorman, J., Hoover, A., Morano, N.C., Ourahmane, A., Srikanth, A., Wang, S., Wu, W., Zhou, T., Andrews, S.F., Kanekiyo, M., Shapiro, L., McDermott, A.B., Kwong, P.D.(2024) Front Immunol 15: 1399960-1399960
- PubMed: 38873606
- DOI: https://doi.org/10.3389/fimmu.2024.1399960
- Primary Citation of Related Structures:
8VUE, 8VUZ, 8VVB - PubMed Abstract:
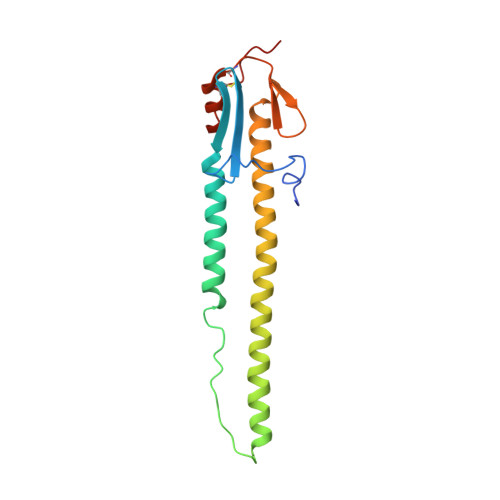
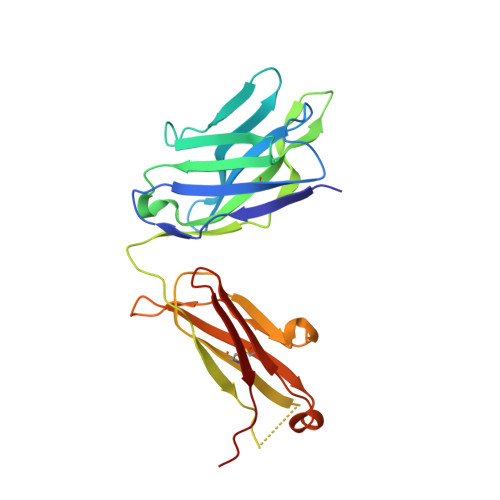
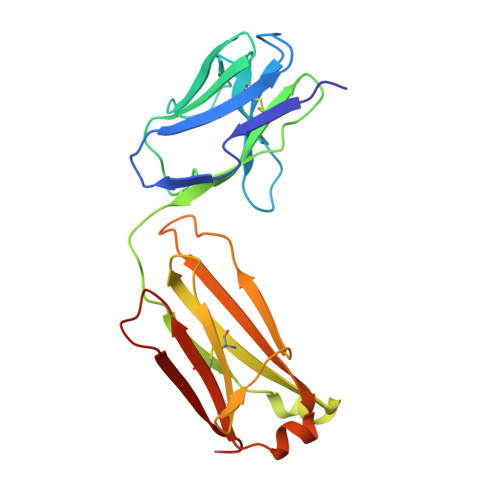
The VH6-1 class of antibodies includes some of the broadest and most potent antibodies that neutralize influenza A virus. Here, we elicit and isolate anti-idiotype antibodies against germline versions of VH6-1 antibodies, use these to sort human leukocytes, and isolate a new VH6-1-class member, antibody L5A7, which potently neutralized diverse group 1 and group 2 influenza A strains. While its heavy chain derived from the canonical IGHV6-1 heavy chain gene used by the class, L5A7 utilized a light chain gene, IGKV1-9, which had not been previously observed in other VH6-1-class antibodies. The cryo-EM structure of L5A7 in complex with Indonesia 2005 hemagglutinin revealed a nearly identical binding mode to other VH6-1-class members. The structure of L5A7 bound to the isolating anti-idiotype antibody, 28H6E11, revealed a shared surface for binding anti-idiotype and hemagglutinin that included two critical L5A7 regions: an FG motif in the third heavy chain-complementary determining region (CDR H3) and the CDR L1 loop. Surprisingly, the chemistries of L5A7 interactions with hemagglutinin and with anti-idiotype were substantially different. Overall, we demonstrate anti-idiotype-based isolation of a broad and potent influenza A virus-neutralizing antibody, revealing that anti-idiotypic selection of antibodies can involve features other than chemical mimicry of the target antigen.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, United States.
Organizational Affiliation: