Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer.
Bakkers, M.J.G., Ritschel, T., Tiemessen, M., Dijkman, J., Zuffiano, A.A., Yu, X., van Overveld, D., Le, L., Voorzaat, R., van Haaren, M.M., de Man, M., Tamara, S., van der Fits, L., Zahn, R., Juraszek, J., Langedijk, J.P.M.(2024) Nat Commun 15: 6270-6270
- PubMed: 39054318
- DOI: https://doi.org/10.1038/s41467-024-50659-5
- Primary Citation of Related Structures:
8VT2, 8VT3 - PubMed Abstract:
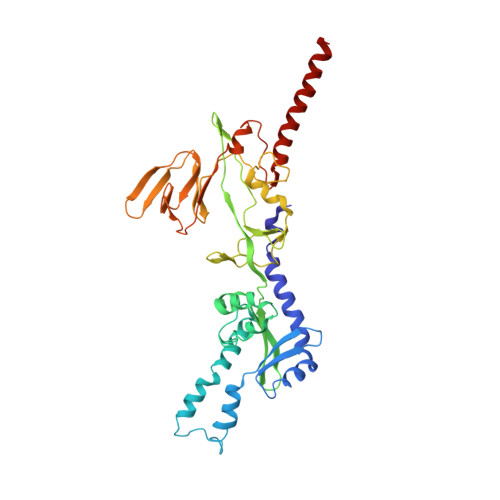
The prefusion conformation of human metapneumovirus fusion protein (hMPV Pre-F) is critical for eliciting the most potent neutralizing antibodies and is the preferred immunogen for an efficacious vaccine against hMPV respiratory infections. Here we show that an additional cleavage event in the F protein allows closure and correct folding of the trimer. We therefore engineered the F protein to undergo double cleavage, which enabled screening for Pre-F stabilizing substitutions at the natively folded protomer interfaces. To identify these substitutions, we developed an AI convolutional classifier that successfully predicts complex polar interactions often overlooked by physics-based methods and visual inspection. The combination of additional processing, stabilization of interface regions and stabilization of the membrane-proximal stem, resulted in a Pre-F protein vaccine candidate without the need for a heterologous trimerization domain that exhibited high expression yields and thermostability. Cryo-EM analysis shows the complete ectodomain structure, including the stem, and a specific interaction of the newly identified cleaved C-terminus with the adjacent protomer. Importantly, the protein induces high and cross-neutralizing antibody responses resulting in near complete protection against hMPV challenge in cotton rats, making the highly stable, double-cleaved hMPV Pre-F trimer an attractive vaccine candidate.
- Janssen Vaccines & Prevention BV, Leiden, The Netherlands.
Organizational Affiliation: