High-resolution single-particle imaging at 100-200 keV with the Gatan Alpine direct electron detector.
Chan, L.M., Courteau, B.J., Maker, A., Wu, M., Basanta, B., Mehmood, H., Bulkley, D., Joyce, D., Lee, B.C., Mick, S., Czarnik, C., Gulati, S., Lander, G.C., Verba, K.A.(2024) J Struct Biol 216: 108108-108108
- PubMed: 38944401
- DOI: https://doi.org/10.1016/j.jsb.2024.108108
- Primary Citation of Related Structures:
8VSU - PubMed Abstract:
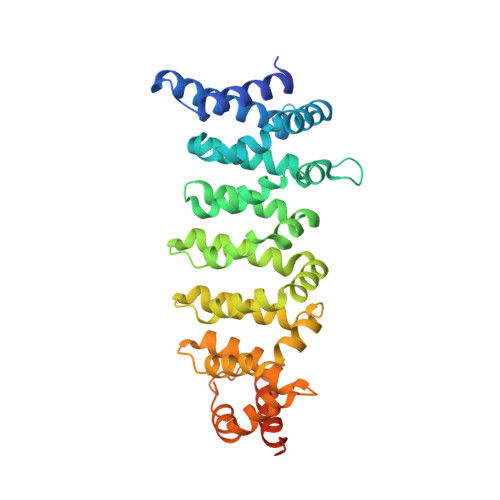
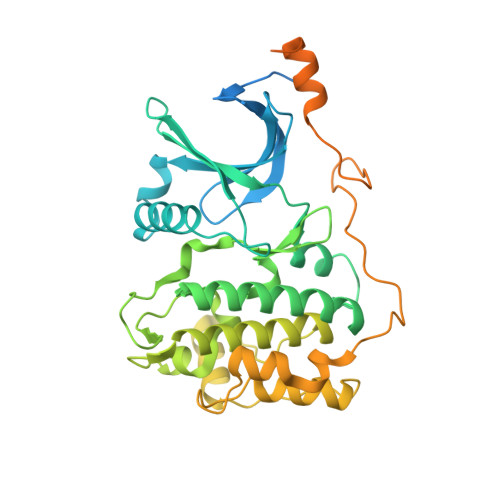
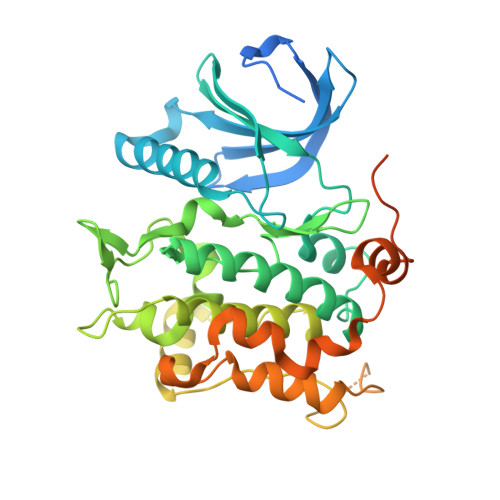
Developments in direct electron detector technology have played a pivotal role in enabling high-resolution structural studies by cryo-EM at 200 and 300 keV. Yet, theory and recent experiments indicate advantages to imaging at 100 keV, energies for which the current detectors have not been optimized. In this study, we evaluated the Gatan Alpine detector, designed for operation at 100 and 200 keV. Compared to the Gatan K3, Alpine demonstrated a significant DQE improvement at these voltages, specifically a ∼ 4-fold improvement at Nyquist at 100 keV. In single-particle cryo-EM experiments, Alpine datasets yielded better than 2 Å resolution reconstructions of apoferritin at 120 and 200 keV on a ThermoFisher Scientific (TFS) Glacios microscope fitted with a non-standard SP-Twin lens. We also achieved a ∼ 3.2 Å resolution reconstruction for a 115 kDa asymmetric protein complex, proving its effectiveness with complex biological samples. In-depth analysis revealed that Alpine reconstructions are comparable to K3 reconstructions at 200 keV, and remarkably, reconstruction from Alpine at 120 keV on a TFS Glacios surpassed all but the 300 keV data from a TFS Titan Krios with GIF/K3. Additionally, we show Alpine's capability for high-resolution data acquisition and screening on lower-end systems by obtaining ∼ 3 Å resolution reconstructions of apoferritin and aldolase at 100 keV and detailed 2D averages of a 55 kDa sample using a side-entry cryo holder. Overall, we show that Gatan Alpine performs well with the standard 200 keV imaging systems and may potentially capture the benefits of lower accelerating voltages, possibly bringing smaller sized particles within the scope of cryo-EM.
- Department of Cellular and Molecular Pharmacology, University of California, San Francisco, CA 94158, United States.
Organizational Affiliation: