Ternary structure of 14-3-3 sigma, ARAF phosphopeptide (pS214) and compound 79 (1124379)
Vickery, H.R., Virta, J.M., Pennings, M., Konstantinidou, M., van den Oetelaar, M., Neitz, R.J., Ottmann, C., Brunsveld, L., Arkin, M.R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 14-3-3 protein sigma | 236 | Homo sapiens | Mutation(s): 0 Gene Names: SFN, HME1 | 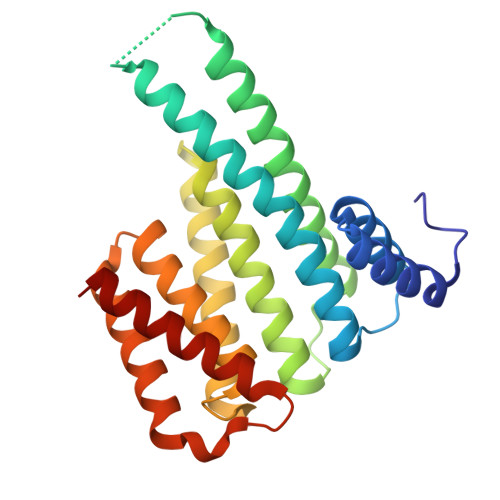 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P31947 (Homo sapiens) Explore P31947 Go to UniProtKB: P31947 | |||||
PHAROS: P31947 GTEx: ENSG00000175793 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P31947 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine/threonine-protein kinase A-Raf phosphopeptide | B [auth P] | 12 | Homo sapiens | Mutation(s): 0 EC: 2.7.11.1 | 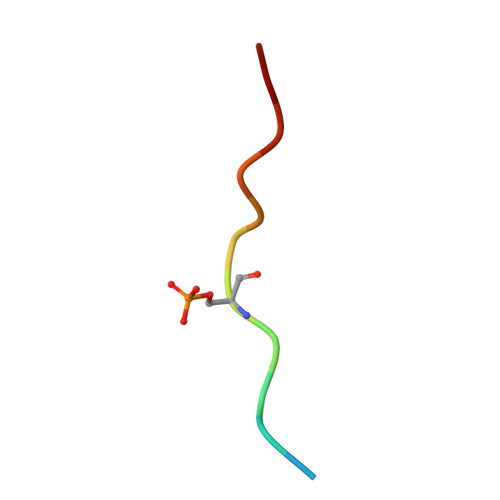 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P10398 (Homo sapiens) Explore P10398 Go to UniProtKB: P10398 | |||||
PHAROS: P10398 GTEx: ENSG00000078061 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10398 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| WQT (Subject of Investigation/LOI) Query on WQT | G [auth A] | 2-chloranyl-1-[8-(4-iodophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]ethanone C14 H16 Cl I N2 O4 S GCSBMYJFFNJXJZ-UHFFFAOYSA-N |  | ||
| CL Query on CL | C [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | D [auth A], E [auth A], F [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | B [auth P] | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.576 | α = 90 |
| b = 150.225 | β = 90 |
| c = 76.816 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PDB-REDO | refinement |
| autoPROC | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM147696 |