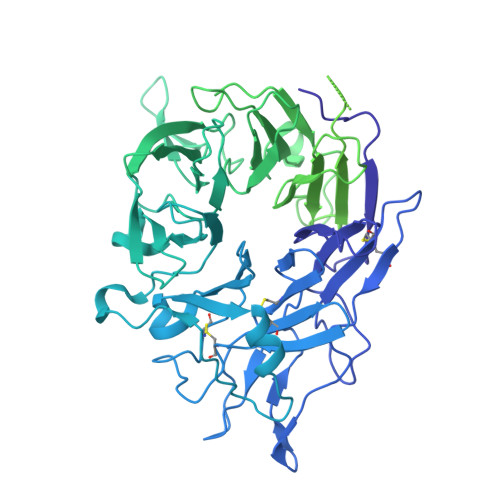
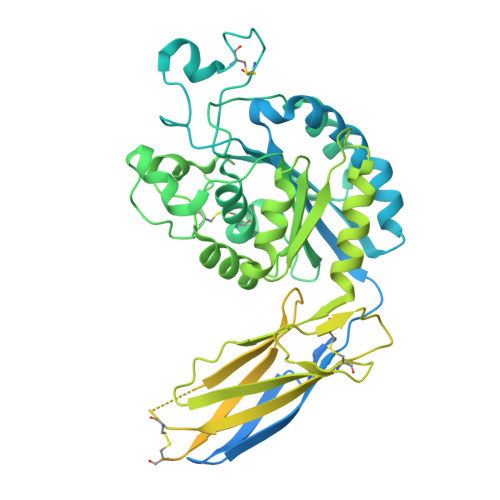
Dynamic allostery drives autocrine and paracrine TGF-beta signaling.
Jin, M., Seed, R.I., Cai, G., Shing, T., Wang, L., Ito, S., Cormier, A., Wankowicz, S.A., Jespersen, J.M., Baron, J.L., Carey, N.D., Campbell, M.G., Yu, Z., Tang, P.K., Cossio, P., Wen, W., Lou, J., Marks, J., Nishimura, S.L., Cheng, Y.(2024) Cell 187: 6200
- PubMed: 39288764
- DOI: https://doi.org/10.1016/j.cell.2024.08.036
- Primary Citation of Related Structures:
8VS6, 8VSB, 8VSC, 8VSD - PubMed Abstract:
TGF-β, essential for development and immunity, is expressed as a latent complex (L-TGF-β) non-covalently associated with its prodomain and presented on immune cell surfaces by covalent association with GARP. Binding to integrin αvβ8 activates L-TGF-β1/GARP. The dogma is that mature TGF-β must physically dissociate from L-TGF-β1 for signaling to occur. Our previous studies discovered that αvβ8-mediated TGF-β autocrine signaling can occur without TGF-β1 release from its latent form. Here, we show that mice engineered to express TGF-β1 that cannot release from L-TGF-β1 survive without early lethal tissue inflammation, unlike those with TGF-β1 deficiency. Combining cryogenic electron microscopy with cell-based assays, we reveal a dynamic allosteric mechanism of autocrine TGF-β1 signaling without release where αvβ8 binding redistributes the intrinsic flexibility of L-TGF-β1 to expose TGF-β1 to its receptors. Dynamic allostery explains the TGF-β3 latency/activation mechanism and why TGF-β3 functions distinctly from TGF-β1, suggesting that it broadly applies to other flexible cell surface receptor/ligand systems.
- Department of Biochemistry and Biophysics, University of California, San Francisco (UCSF), San Francisco, CA, USA.
Organizational Affiliation: