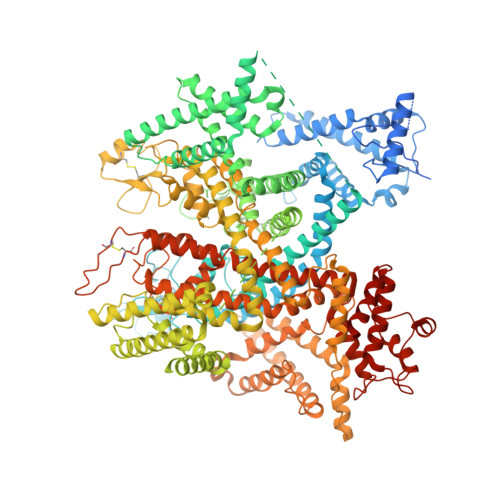
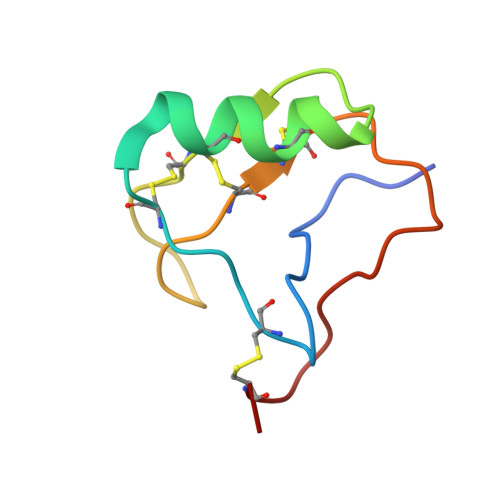
Scorpion alpha-toxin Lqh alpha IT specifically interacts with a glycan at the pore domain of voltage-gated sodium channels.
Phulera, S., Dickson, C.J., Schwalen, C.J., Khoshouei, M., Cassell, S.J., Sun, Y., Condos, T., Whicher, J., Weihofen, W.A.(2024) Structure 32: 1611-1620.e4
- PubMed: 39181123
- DOI: https://doi.org/10.1016/j.str.2024.07.021
- Primary Citation of Related Structures:
8VQC - PubMed Abstract:
Voltage-gated sodium (Nav) channels sense membrane potential and drive cellular electrical activity. The deathstalker scorpion α-toxin LqhαIT exerts a strong action potential prolonging effect on Nav channels. To elucidate the mechanism of action of LqhαIT, we determined a 3.9 Å cryoelectron microscopy (cryo-EM) structure of LqhαIT in complex with the Nav channel from Periplaneta americana (NavPas). We found that LqhαIT binds to voltage sensor domain 4 and traps it in an "S4 down" conformation. The functionally essential C-terminal epitope of LqhαIT forms an extensive interface with the glycan scaffold linked to Asn330 of NavPas that augments a small protein-protein interface between NavPas and LqhαIT. A combination of molecular dynamics simulations, structural comparisons, and prior mutagenesis experiments demonstrates the functional importance of this toxin-glycan interaction. These findings establish a structural basis for the specificity achieved by scorpion α-toxins and reveal the conserved glycan as an essential component of the toxin-binding epitope.
- Discovery Sciences, Novartis Biomedical Research, 250 Massachusetts Avenue, Cambridge, MA 02139, USA.
Organizational Affiliation: