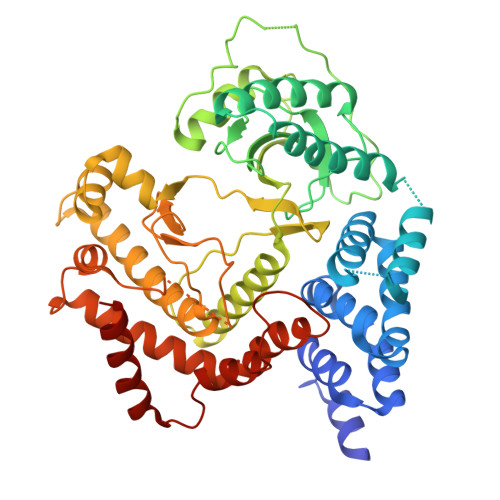
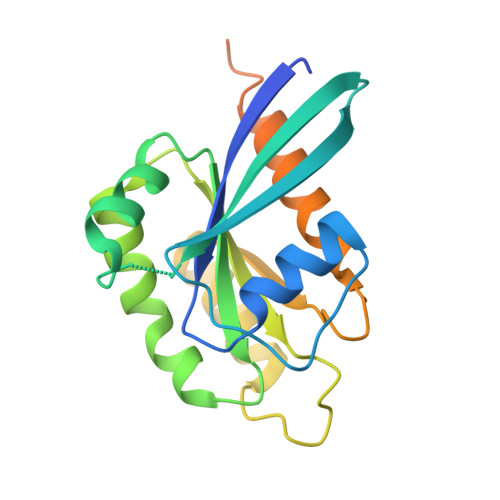
Cryptosporidium PI(4)K inhibitor EDI048 is a gut-restricted parasiticidal agent to treat paediatric enteric cryptosporidiosis.
Manjunatha, U.H., Lakshminarayana, S.B., Jumani, R.S., Chao, A.T., Young, J.M., Gable, J.E., Knapp, M., Hanna, I., Galarneau, J.R., Cantwell, J., Kulkarni, U., Turner, M., Lu, P., Darrell, K.H., Watson, L.C., Chan, K., Patra, D., Mamo, M., Luu, C., Cuellar, C., Shaul, J., Xiao, L., Chen, Y.B., Carney, S.K., Lakshman, J., Osborne, C.S., Zambriski, J.A., Aziz, N., Sarko, C., Diagana, T.T.(2024) Nat Microbiol 9: 2817-2835
- PubMed: 39379634
- DOI: https://doi.org/10.1038/s41564-024-01810-x
- Primary Citation of Related Structures:
8VOF - PubMed Abstract:
Diarrhoeal disease caused by Cryptosporidium is a major cause of morbidity and mortality in young and malnourished children from low- and middle-income countries, with no vaccine or effective treatment. Here we describe the discovery of EDI048, a Cryptosporidium PI(4)K inhibitor, designed to be active at the infection site in the gastrointestinal tract and undergo rapid metabolism in the liver. By using mutational analysis and crystal structure, we show that EDI048 binds to highly conserved amino acid residues in the ATP-binding site. EDI048 is orally efficacious in an immunocompromised mouse model despite negligible circulating concentrations, thus demonstrating that gastrointestinal exposure is necessary and sufficient for efficacy. In neonatal calves, a clinical model of cryptosporidiosis, EDI048 treatment resulted in rapid resolution of diarrhoea and significant reduction in faecal oocyst shedding. Safety and pharmacological studies demonstrated predictable metabolism and low systemic exposure of EDI048, providing a substantial safety margin required for a paediatric indication. EDI048 is a promising clinical candidate for the treatment of life-threatening paediatric cryptosporidiosis.
- Global Health, Biomedical Research, Novartis, Emeryville, CA, USA. manjunatha.ujjini@novartis.com.
Organizational Affiliation: