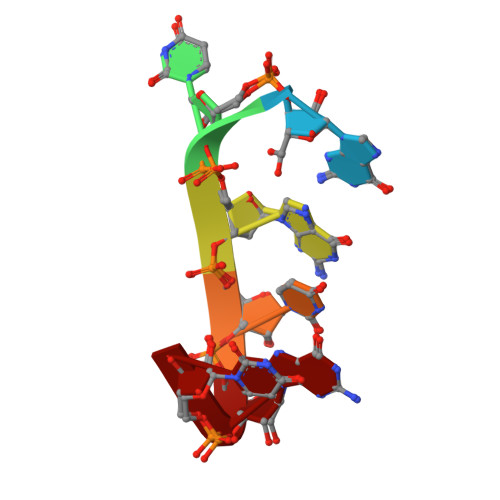
Self-assembly and condensation of intermolecular poly(UG) RNA quadruplexes.
Roschdi, S., Montemayor, E.J., Vivek, R., Bingman, C.A., Butcher, S.E.(2024) Nucleic Acids Res 52: 12582-12591
- PubMed: 39373474
- DOI: https://doi.org/10.1093/nar/gkae870
- Primary Citation of Related Structures:
8VJT - PubMed Abstract:
Poly(UG) or 'pUG' dinucleotide repeats are highly abundant sequences in eukaryotic RNAs. In Caenorhabditis elegans, pUGs are added to RNA 3' ends to direct gene silencing within Mutator foci, a germ granule condensate. Here, we show that pUG RNAs efficiently self-assemble into gel condensates through quadruplex (G4) interactions. Short pUG sequences form right-handed intermolecular G4s (pUG G4s), while longer pUGs form left-handed intramolecular G4s (pUG folds). We determined a 1.05 Å crystal structure of an intermolecular pUG G4, which reveals an eight stranded G4 dimer involving 48 nucleotides, 7 different G and U quartet conformations, 7 coordinated potassium ions, 8 sodium ions and a buried water molecule. A comparison of the intermolecular pUG G4 and intramolecular pUG fold structures provides insights into the molecular basis for G4 handedness and illustrates how a simple dinucleotide repeat sequence can form complex structures with diverse topologies.
- Department of Biochemistry, University of Wisconsin-Madison, Madison, WI 53706, USA.
Organizational Affiliation: