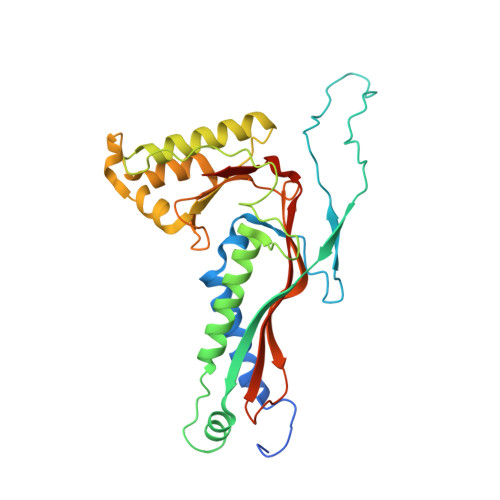
Myxococcus xanthus encapsulin cargo protein EncD is a flavin-binding protein with ferric reductase activity.
Eren, E., Watts, N.R., Conway, J.F., Wingfield, P.T.(2024) Proc Natl Acad Sci U S A 121: e2400426121-e2400426121
- PubMed: 38748579
- DOI: https://doi.org/10.1073/pnas.2400426121
- Primary Citation of Related Structures:
8VJN, 8VJO - PubMed Abstract:
Encapsulins are protein nanocompartments that regulate cellular metabolism in several bacteria and archaea. Myxococcus xanthus encapsulins protect the bacterial cells against oxidative stress by sequestering cytosolic iron. These encapsulins are formed by the shell protein EncA and three cargo proteins: EncB, EncC, and EncD. EncB and EncC form rotationally symmetric decamers with ferroxidase centers (FOCs) that oxidize Fe +2 to Fe +3 for iron storage in mineral form. However, the structure and function of the third cargo protein, EncD, have yet to be determined. Here, we report the x-ray crystal structure of EncD in complex with flavin mononucleotide. EncD forms an α-helical hairpin arranged as an antiparallel dimer, but unlike other flavin-binding proteins, it has no β-sheet, showing that EncD and its homologs represent a unique class of bacterial flavin-binding proteins. The cryo-EM structure of EncA-EncD encapsulins confirms that EncD binds to the interior of the EncA shell via its C-terminal targeting peptide. With only 100 amino acids, the EncD α-helical dimer forms the smallest flavin-binding domain observed to date. Unlike EncB and EncC, EncD lacks a FOC, and our biochemical results show that EncD instead is a NAD(P)H-dependent ferric reductase, indicating that the M. xanthus encapsulins act as an integrated system for iron homeostasis. Overall, this work contributes to our understanding of bacterial metabolism and could lead to the development of technologies for iron biomineralization and the production of iron-containing materials for the treatment of various diseases associated with oxidative stress.
- Protein Expression Laboratory, National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH, Bethesda, MD 20892.
Organizational Affiliation: