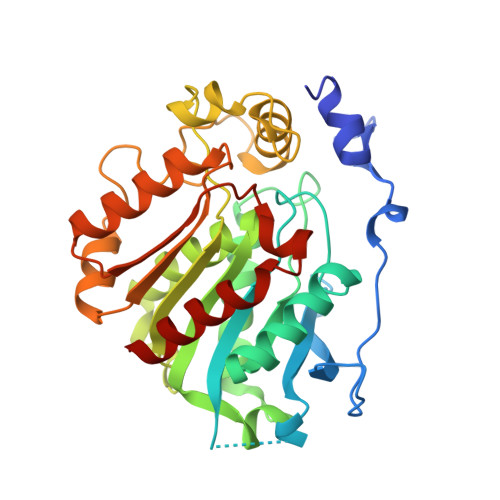
Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Palayam, M., Yan, L., Nagalakshmi, U., Gilio, A.K., Cornu, D., Boyer, F.D., Dinesh-Kumar, S.P., Shabek, N.(2024) Nat Commun 15: 6500-6500
- PubMed: 39090154
- DOI: https://doi.org/10.1038/s41467-024-50928-3
- Primary Citation of Related Structures:
8VCA, 8VCD, 8VCE - PubMed Abstract:
Phytohormone levels are regulated through specialized enzymes, participating not only in their biosynthesis but also in post-signaling processes for signal inactivation and cue depletion. Arabidopsis thaliana (At) carboxylesterase 15 (CXE15) and carboxylesterase 20 (CXE20) have been shown to deplete strigolactones (SLs) that coordinate various growth and developmental processes and function as signaling molecules in the rhizosphere. Here, we elucidate the X-ray crystal structures of AtCXE15 (both apo and SL intermediate bound) and AtCXE20, revealing insights into the mechanisms of SL binding and catabolism. The N-terminal regions of CXE15 and CXE20 exhibit distinct secondary structures, with CXE15 characterized by an alpha helix and CXE20 by an alpha/beta fold. These structural differences play pivotal roles in regulating variable SL hydrolysis rates. Our findings, both in vitro and in planta, indicate that a transition of the N-terminal helix domain of CXE15 between open and closed forms facilitates robust SL hydrolysis. The results not only illuminate the distinctive process of phytohormone breakdown but also uncover a molecular architecture and mode of plasticity within a specific class of carboxylesterases.
- Department of Plant Biology, College of Biological Sciences, University of California-Davis, Davis, CA, USA.
Organizational Affiliation: