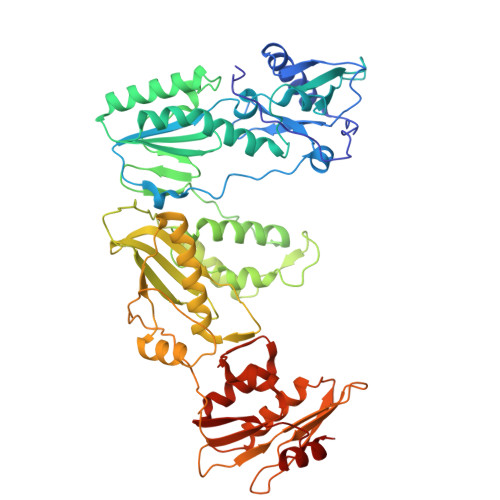
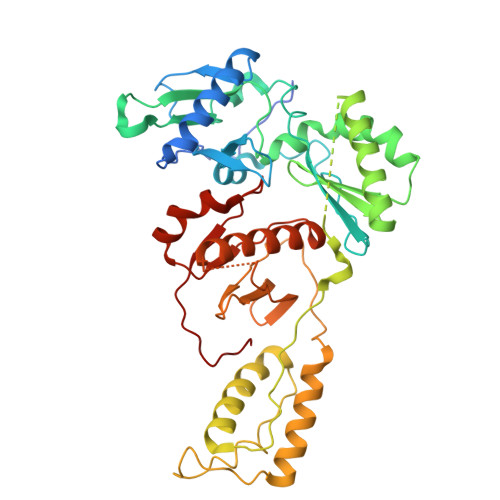
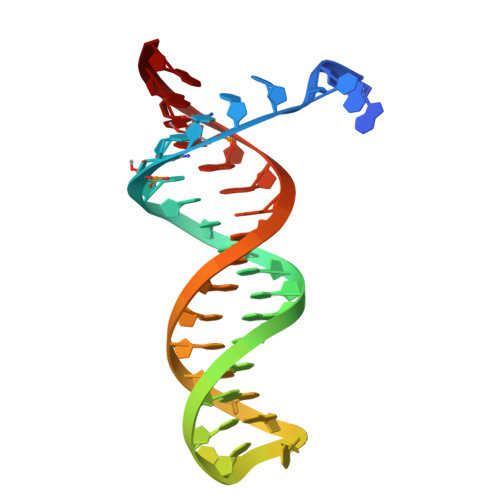
Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Vergara, S., Zhou, X., Santiago, U., Alaoui-El-Azher, M., Conway, J.F., Sluis-Cremer, N., Calero, G.(2024) Nat Commun 15: 10553-10553
- PubMed: 39632888
- DOI: https://doi.org/10.1038/s41467-024-54618-y
- Primary Citation of Related Structures:
8VB6, 8VB7, 8VB8, 8VB9, 8VBC, 8VBD, 8VBE, 8VBF, 8VBG, 8VBH, 8VBI - PubMed Abstract:
Reverse transcription of the retroviral RNA genome into DNA is an integral step during HIV-1 replication. Despite a wealth of structural information on reverse transcriptase (RT), we lack insight into the intermediate states of DNA synthesis. Using catalytically active substrates, and a blot/diffusion cryo-electron microscopy approach, we capture 11 structures encompassing reactant, intermediate and product states of dATP addition by RT at 2.2 to 3.0 Å resolution. In the reactant state, dATP binding to RT-template/primer involves a single Mg 2+ (site B) inducing formation of a negatively charged pocket where a second floating Mg 2+ can bind (site A). During the intermediate state, the α-phosphate oxygen from a previously unobserved dATP conformer aligns with site A Mg 2+ and the primer 3'-OH for nucleophilic attack. The product state, comprises two substrate conformations including an incorporated dAMP with the pyrophosphate leaving group coordinated by metal B and stabilized through H-bonds. Moreover, K220 mutants significantly impact the rate of dNTP incorporation by RT and HIV-1 replication capacity. This work sheds light into the dynamic components of a reaction that is central to HIV-1 replication.
- Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA.
Organizational Affiliation: