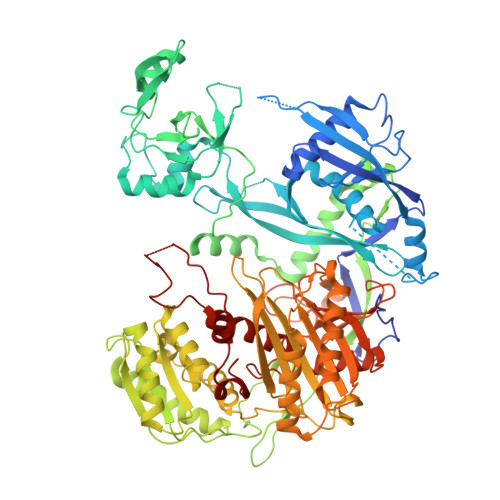
Target cleavage and gene silencing by Argonautes with cityRNAs.
Zhang, H., Sim, G., Kehling, A.C., Adhav, V.A., Savidge, A., Pastore, B., Tang, W., Nakanishi, K.(2024) Cell Rep 43: 114806-114806
- PubMed: 39368090
- DOI: https://doi.org/10.1016/j.celrep.2024.114806
- Primary Citation of Related Structures:
8VAJ - PubMed Abstract:
TinyRNAs (tyRNAs) are ≤17-nt guide RNAs associated with Argonaute proteins (AGOs), and certain 14-nt cleavage-inducing tyRNAs (cityRNAs) catalytically activate human Argonaute3 (AGO3). We present the crystal structure of AGO3 in complex with a cityRNA, 14-nt miR-20a, and its complementary target, revealing a different trajectory for the guide-target duplex from that of its ∼22-nt microRNA-associated AGO counterpart. cityRNA-loaded Argonaute2 (AGO2) and AGO3 enhance their endonuclease activity when the immediate 5' upstream region of the tyRNA target site (UTy) includes sequences with low affinity for AGO. We propose a model where cityRNA-loaded AGO2 and AGO3 efficiently cleave fully complementary tyRNA target sites unless they directly recognize the UTy. To investigate their gene silencing, we devised systems for loading endogenous AGOs with specific tyRNAs and demonstrated that, unlike microRNAs, cityRNA-mediated silencing heavily relies on target cleavage. Our study uncovered that AGO exploits cityRNAs for target recognition differently from microRNAs and alters gene silencing.
- Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA.
Organizational Affiliation: