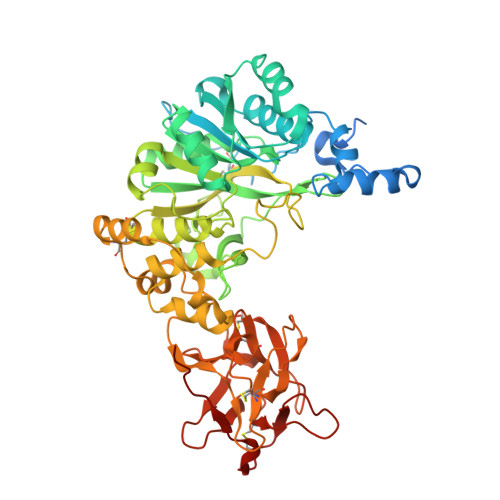
An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Collette, A.M., Hassan, S.A., Schmidt, S.I., Lara, A.J., Yang, W., Samara, N.L.(2024) Sci Adv 10: eadj8829-eadj8829
- PubMed: 38416819
- DOI: https://doi.org/10.1126/sciadv.adj8829
- Primary Citation of Related Structures:
8V9Q - PubMed Abstract:
N-acetylgalactosaminyl-transferases (GalNAc-Ts) initiate mucin-type O-glycosylation, an abundant and complex posttranslational modification that regulates host-microbe interactions, tissue development, and metabolism. GalNAc-Ts contain a lectin domain consisting of three homologous repeats (α, β, and γ), where α and β can potentially interact with O-GalNAc on substrates to enhance activity toward a nearby acceptor Thr/Ser. The ubiquitous isoenzyme GalNAc-T1 modulates heart development, immunity, and SARS-CoV-2 infectivity, but its substrates are largely unknown. Here, we show that both α and β in GalNAc-T1 uniquely orchestrate the O-glycosylation of various glycopeptide substrates. The α repeat directs O-glycosylation to acceptor sites carboxyl-terminal to an existing GalNAc, while the β repeat directs O-glycosylation to amino-terminal sites. In addition, GalNAc-T1 incorporates α and β into various substrate binding modes to cooperatively increase the specificity toward an acceptor site located between two existing O-glycans. Our studies highlight a unique mechanism by which dual lectin repeats expand substrate specificity and provide crucial information for identifying the biological substrates of GalNAc-T1.
- Structural Biochemistry Unit, NIDCR, NIH, Bethesda, MD 20892, USA.
Organizational Affiliation: