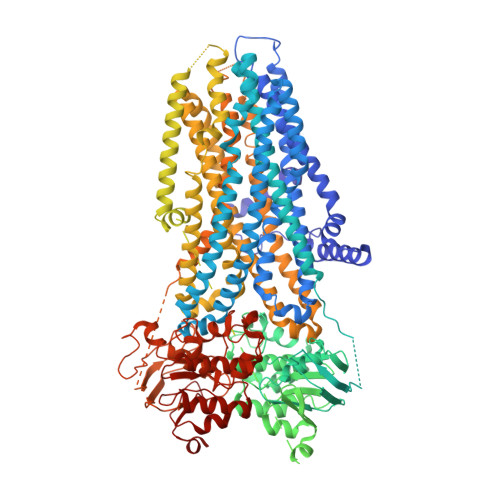
Allosteric inhibition of CFTR gating by CFTRinh-172 binding in the pore.
Gao, X., Yeh, H.I., Yang, Z., Fan, C., Jiang, F., Howard, R.J., Lindahl, E., Kappes, J.C., Hwang, T.C.(2024) Nat Commun 15: 6668-6668
- PubMed: 39107303
- DOI: https://doi.org/10.1038/s41467-024-50641-1
- Primary Citation of Related Structures:
8V7Z, 8V81 - PubMed Abstract:
Loss-of-function mutations of the CFTR gene cause the life-shortening genetic disease cystic fibrosis (CF), whereas overactivity of CFTR may lead to secretory diarrhea and polycystic kidney disease. While effective drugs targeting the CFTR protein have been developed for the treatment of CF, little progress has been made for diseases caused by hyper-activated CFTR. Here, we solve the cryo-EM structure of CFTR in complex with CFTRinh-172 (Inh-172), a CFTR gating inhibitor with promising potency and efficacy. We find that Inh-172 binds inside the pore of CFTR, interacting with amino acid residues from transmembrane segments (TMs) 1, 6, 8, 9, and 12 through mostly hydrophobic interactions and a salt bridge. Substitution of these residues lowers the apparent affinity of Inh-172. The inhibitor-bound structure reveals re-orientations of the extracellular segment of TMs 1, 8, and 12, supporting an allosteric modulation mechanism involving post-binding conformational changes. This allosteric inhibitory mechanism readily explains our observations that pig CFTR, which preserves all the amino acid residues involved in Inh-172 binding, exhibits a much-reduced sensitivity to Inh-172 and that the apparent affinity of Inh-172 is altered by the CF drug ivacaftor (i.e., VX-770) which enhances CFTR's activity through binding to a site also comprising TM8.
- Dalton Cardiovascular Research Center, University of Missouri-Columbia, Columbia, MO, 65211, USA. xgdz2@missouri.edu.
Organizational Affiliation: