RCK1-RCK2 double mutant of human Slo1 in presence of EDTA - resting VSD
Pal, K., Kallure, G.S., Chowdhury, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Calcium-activated potassium channel subunit alpha-1 | 1,065 | Homo sapiens | Mutation(s): 6 Gene Names: KCNMA1, KCNMA, SLO Membrane Entity: Yes | 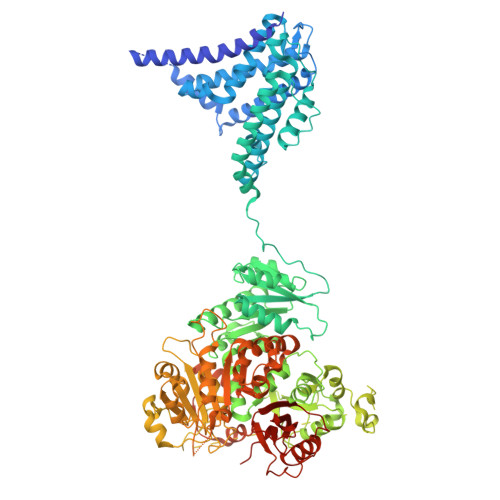 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q12791 (Homo sapiens) Explore Q12791 Go to UniProtKB: Q12791 | |||||
PHAROS: Q12791 GTEx: ENSG00000156113 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12791 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AJP (Subject of Investigation/LOI) Query on AJP | AB [auth C] BB [auth C] CB [auth C] DB [auth C] EB [auth C] | Digitonin C56 H92 O29 UVYVLBIGDKGWPX-KUAJCENISA-N |  | ||
| POV (Subject of Investigation/LOI) Query on POV | BA [auth B] CA [auth B] DA [auth B] EA [auth B] FA [auth B] | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate C42 H82 N O8 P WTJKGGKOPKCXLL-PFDVCBLKSA-N |  | ||
| CLR (Subject of Investigation/LOI) Query on CLR | HA [auth B], OB [auth D], Q [auth A], YA [auth C] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| K (Subject of Investigation/LOI) Query on K | AA [auth A] E [auth A] F [auth A] G [auth A] H [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R01GM145719 |