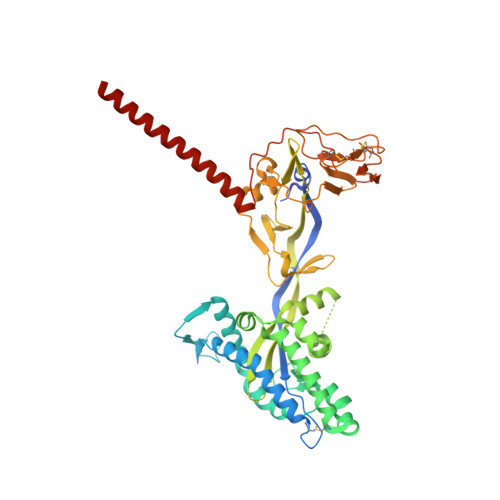
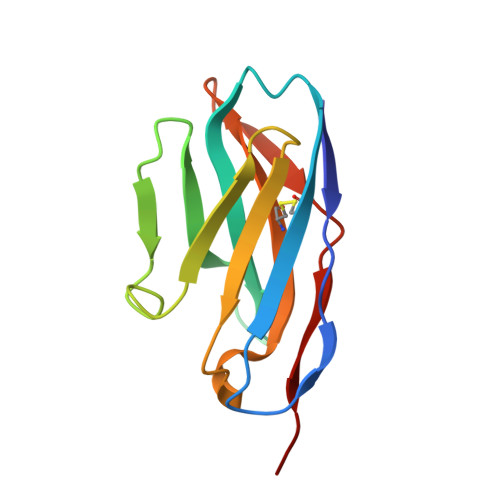
Universal paramyxovirus vaccine design by stabilizing regions involved in structural transformation of the fusion protein.
Langedijk, J.P.M., Cox, F., Johnson, N.V., van Overveld, D., Le, L., van den Hoogen, W., Voorzaat, R., Zahn, R., van der Fits, L., Juraszek, J., McLellan, J.S., Bakkers, M.J.G.(2024) Nat Commun 15: 4629-4629
- PubMed: 38821950
- DOI: https://doi.org/10.1038/s41467-024-48059-w
- Primary Citation of Related Structures:
8V5A - PubMed Abstract:
The Paramyxoviridae family encompasses medically significant RNA viruses, including human respiroviruses 1 and 3 (RV1, RV3), and zoonotic pathogens like Nipah virus (NiV). RV3, previously known as parainfluenza type 3, for which no vaccines or antivirals have been approved, causes respiratory tract infections in vulnerable populations. The RV3 fusion (F) protein is inherently metastable and will likely require prefusion (preF) stabilization for vaccine effectiveness. Here we used structure-based design to stabilize regions involved in structural transformation to generate a preF protein vaccine antigen with high expression and stability, and which, by stabilizing the coiled-coil stem region, does not require a heterologous trimerization domain. The preF candidate induces strong neutralizing antibody responses in both female naïve and pre-exposed mice and provides protection in a cotton rat challenge model (female). Despite the evolutionary distance of paramyxovirus F proteins, their structural transformation and local regions of instability are conserved, which allows successful transfer of stabilizing substitutions to the distant preF proteins of RV1 and NiV. This work presents a successful vaccine antigen design for RV3 and provides a toolbox for future paramyxovirus vaccine design and pandemic preparedness.
- Janssen Vaccines & Prevention BV, Leiden, The Netherlands.
Organizational Affiliation: