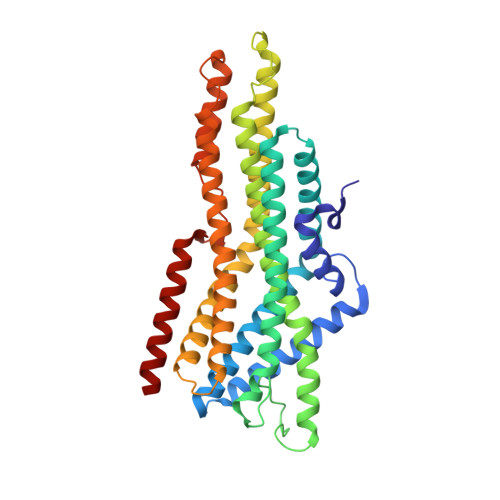
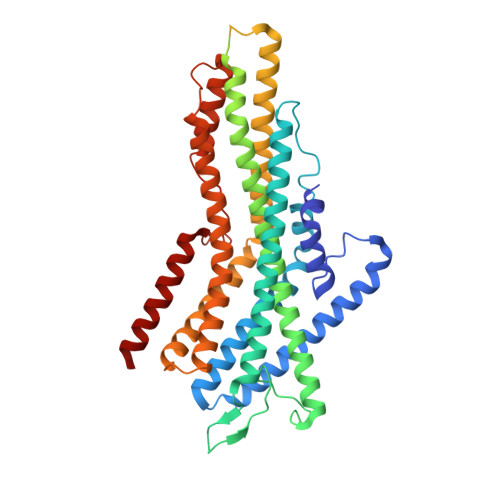
Structural basis of odor sensing by insect heteromeric odorant receptors.
Zhao, J., Chen, A.Q., Ryu, J., Del Marmol, J.(2024) Science 384: 1460-1467
- PubMed: 38870275
- DOI: https://doi.org/10.1126/science.adn6384
- Primary Citation of Related Structures:
8V00, 8V02, 8V3C, 8V3D - PubMed Abstract:
Most insects, including human-targeting mosquitoes, detect odors through odorant-activated ion channel complexes consisting of a divergent odorant-binding subunit (OR) and a conserved co-receptor subunit (Orco). As a basis for understanding how odorants activate these heteromeric receptors, we report here cryo-electron microscopy structures of two different heteromeric odorant receptor complexes containing ORs from disease-vector mosquitos Aedes aegypti or Anopheles gambiae . These structures reveal an unexpected stoichiometry of one OR to three Orco subunits. Comparison of structures in odorant-bound and unbound states indicates that odorant binding to the sole OR subunit is sufficient to open the channel pore, suggesting a mechanism of OR activation and a conceptual framework for understanding evolution of insect odorant receptor sensitivity.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: