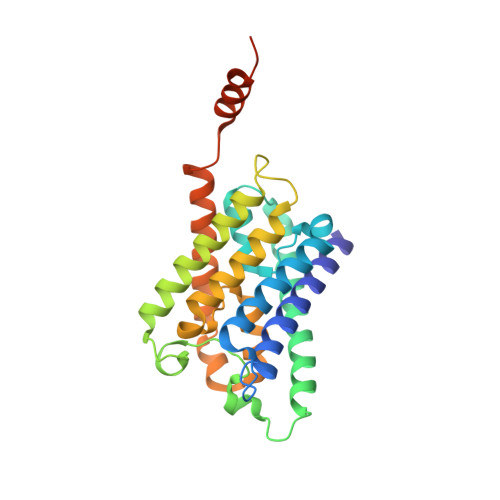
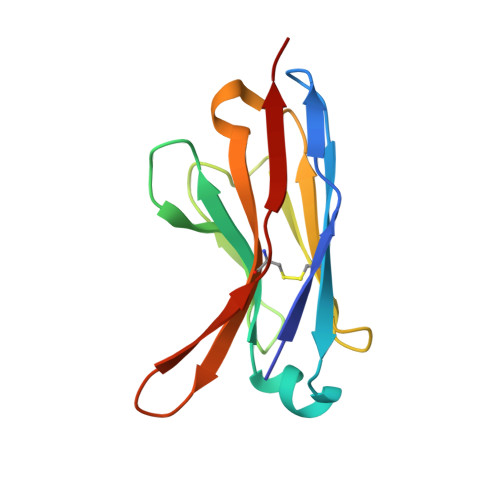
Grafting the ALFA tag for structural studies of aquaporin Z.
Stover, L., Bahramimoghaddam, H., Wang, L., Schrecke, S., Yadav, G.P., Zhou, M., Laganowsky, A.(2024) J Struct Biol X 9: 100097-100097
- PubMed: 38361954
- DOI: https://doi.org/10.1016/j.yjsbx.2024.100097
- Primary Citation of Related Structures:
8UY6 - PubMed Abstract:
Aquaporin Z (AqpZ), a bacterial water channel, forms a tetrameric complex and, like many other membrane proteins, activity is regulated by lipids. Various methods have been developed to facilitate structure determination of membrane proteins, such as the use of antibodies. Here, we graft onto AqpZ the ALFA tag (AqpZ-ALFA), an alpha helical epitope, to make use of the high-affinity anti-ALFA nanobody (nB). Native mass spectrometry reveals the AqpZ-ALFA fusion forms a stable, 1:1 complex with nB. Single-particle cryogenic electron microscopy studies reveal the octameric (AqpZ-ALFA) 4 (nB) 4 complex forms a dimeric assembly and the structure was determined to 1.9 Å resolution. Dimerization of the octamer is mediated through stacking of the symmetrically bound nBs. Tube-like density is also observed, revealing a potential cardiolipin binding site. Grafting of the ALFA tag, or other epitope, along with binding and association of nBs to promote larger complexes will have applications in structural studies and protein engineering.
- Department of Chemistry, Texas A&M University, College Station, TX 77843, United States.
Organizational Affiliation: