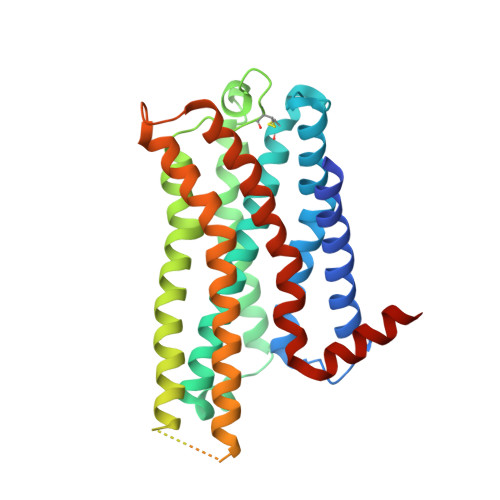
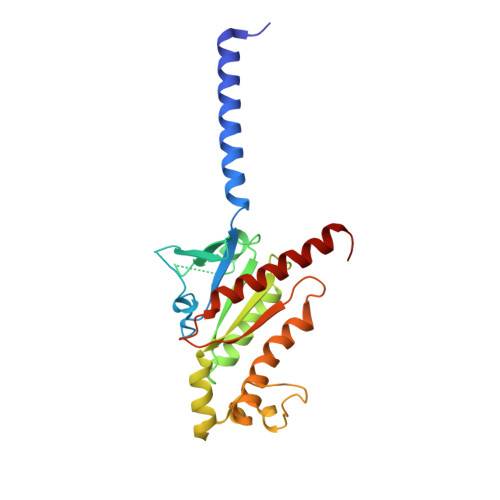
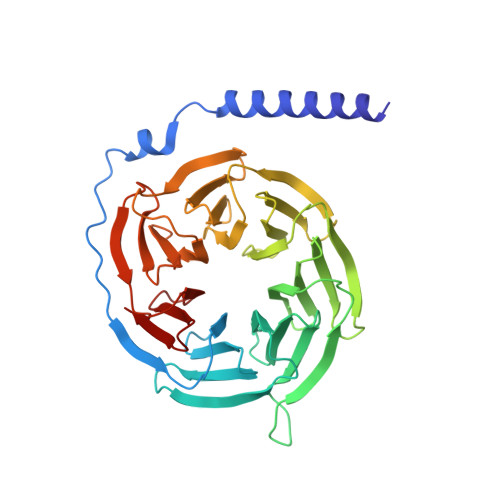
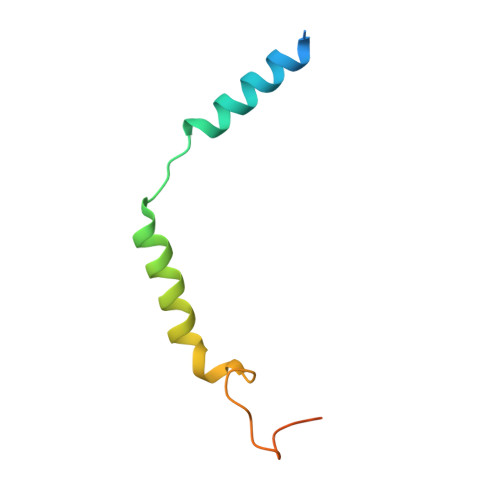
AlphaFold2 structures guide prospective ligand discovery.
Lyu, J., Kapolka, N., Gumpper, R., Alon, A., Wang, L., Jain, M.K., Barros-Alvarez, X., Sakamoto, K., Kim, Y., DiBerto, J., Kim, K., Glenn, I.S., Tummino, T.A., Huang, S., Irwin, J.J., Tarkhanova, O.O., Moroz, Y., Skiniotis, G., Kruse, A.C., Shoichet, B.K., Roth, B.L.(2024) Science 384: eadn6354-eadn6354
- PubMed: 38753765
- DOI: https://doi.org/10.1126/science.adn6354
- Primary Citation of Related Structures:
8UWL, 8V6U - PubMed Abstract:
AlphaFold2 (AF2) models have had wide impact, but they have had mixed success in retrospective ligand recognition. We prospectively docked large libraries against unrefined AF2 models of the σ2 and 5-HT2A receptors, testing hundreds of new molecules and comparing results to docking against the experimental structures. Hit rates were high and similar for the experimental and the AF2 structures, as were affinities. The success of docking against the AF2 models was achieved despite differences in orthosteric residue conformations versus the experimental structures. Determination of the cryo-electron microscopy structure for one of the more potent 5HT2A ligands from the AF2 docking revealed residue accommodations that resembled the AF2 prediction. AF2 models may sample conformations that differ from experimental structures but remain low energy and relevant for ligand discovery, extending the domain of structure-based ligand discovery.
- Department of Pharmaceutical Chemistry, University of California, San Francisco, CA 94158, USA.
Organizational Affiliation: