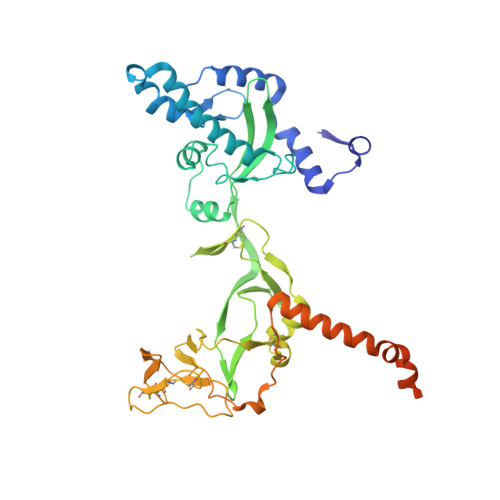
A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Zyla, D.S., Della Marca, R., Niemeyer, G., Zipursky, G., Stearns, K., Leedale, C., Sobolik, E.B., Callaway, H.M., Hariharan, C., Peng, W., Parekh, D., Marcink, T.C., Diaz Avalos, R., Horvat, B., Mathieu, C., Snijder, J., Greninger, A.L., Hastie, K.M., Niewiesk, S., Moscona, A., Porotto, M., Ollmann Saphire, E.(2024) Science 384: eadm8693-eadm8693
- PubMed: 38935733
- DOI: https://doi.org/10.1126/science.adm8693
- Primary Citation of Related Structures:
8UT2, 8UTF, 8UUP, 8UUQ, 9AT8 - PubMed Abstract:
Measles virus (MeV) presents a public health threat that is escalating as vaccine coverage in the general population declines and as populations of immunocompromised individuals, who cannot be vaccinated, increase. There are no approved therapeutics for MeV. Neutralizing antibodies targeting viral fusion are one potential therapeutic approach but have not yet been structurally characterized or advanced to clinical use. We present cryo-electron microscopy (cryo-EM) structures of prefusion F alone [2.1-angstrom (Å) resolution], F complexed with a fusion-inhibitory peptide (2.3-Å resolution), F complexed with the neutralizing and protective monoclonal antibody (mAb) 77 (2.6-Å resolution), and an additional structure of postfusion F (2.7-Å resolution). In vitro assays and examination of additional EM classes show that mAb 77 binds prefusion F, arrests F in an intermediate state, and prevents transition to the postfusion conformation. These structures shed light on antibody-mediated neutralization that involves arrest of fusion proteins in an intermediate state.
- Center for Vaccine Innovation, La Jolla Institute for Immunology, La Jolla, CA 92037, USA.
Organizational Affiliation: