Intracellular Ebola virus nucleocapsid assembly revealed by in situ cryo-electron tomography.
Watanabe, R., Zyla, D., Parekh, D., Hong, C., Jones, Y., Schendel, S.L., Wan, W., Castillon, G., Saphire, E.O.(2024) Cell 187: 5587-5603.e19
- PubMed: 39293445
- DOI: https://doi.org/10.1016/j.cell.2024.08.044
- Primary Citation of Related Structures:
8USN, 8UST - PubMed Abstract:
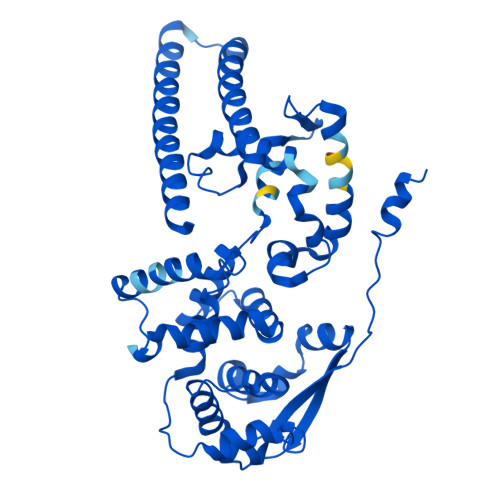
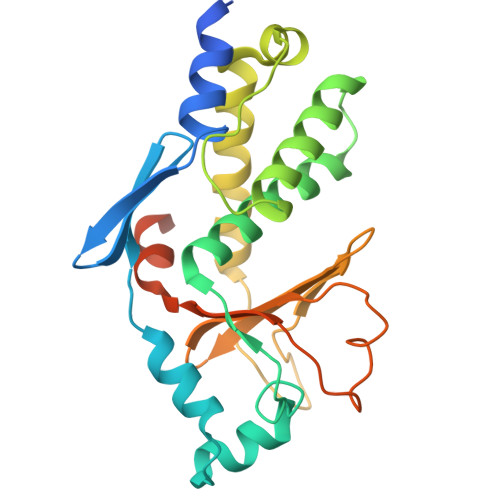
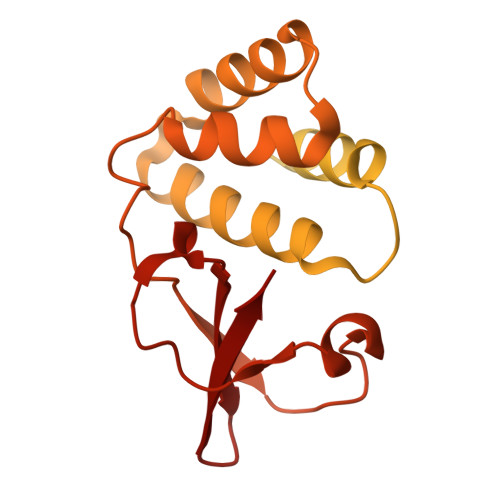
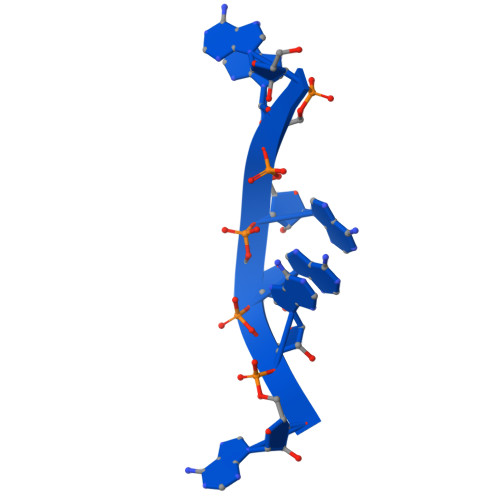
Filoviruses, including the Ebola and Marburg viruses, cause hemorrhagic fevers with up to 90% lethality. The viral nucleocapsid is assembled by polymerization of the nucleoprotein (NP) along the viral genome, together with the viral proteins VP24 and VP35. We employed cryo-electron tomography of cells transfected with viral proteins and infected with model Ebola virus to illuminate assembly intermediates, as well as a 9 Å map of the complete intracellular assembly. This structure reveals a previously unresolved third and outer layer of NP complexed with VP35. The intrinsically disordered region, together with the C-terminal domain of this outer layer of NP, provides the constant width between intracellular nucleocapsid bundles and likely functions as a flexible tether to the viral matrix protein in the virion. A comparison of intracellular nucleocapsids with prior in-virion nucleocapsid structures reveals that the nucleocapsid further condenses vertically in the virion. The interfaces responsible for nucleocapsid assembly are highly conserved and offer targets for broadly effective antivirals.
- Center for Vaccine Innovation, La Jolla Institute for Immunology, La Jolla, CA 92037, USA.
Organizational Affiliation: