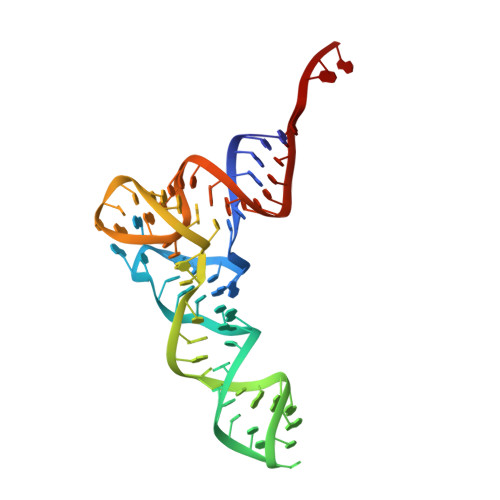
tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Krahn, N., Zhang, J., Melnikov, S.V., Tharp, J.M., Villa, A., Patel, A., Howard, R.J., Gabir, H., Patel, T.R., Stetefeld, J., Puglisi, J., Soll, D.(2024) Nucleic Acids Res 52: 513-524
- PubMed: 38100361
- DOI: https://doi.org/10.1093/nar/gkad1188
- Primary Citation of Related Structures:
8UPT, 8UPY - PubMed Abstract:
Protein translation is orchestrated through tRNA aminoacylation and ribosomal elongation. Among the highly conserved structure of tRNAs, they have distinguishing features which promote interaction with their cognate aminoacyl tRNA synthetase (aaRS). These key features are referred to as identity elements. In our study, we investigated the tRNA:aaRS pair that installs the 22nd amino acid, pyrrolysine (tRNAPyl:PylRS). Pyrrolysyl-tRNA synthetases (PylRSs) are naturally encoded in some archaeal and bacterial genomes to acylate tRNAPyl with pyrrolysine. Their large amino acid binding pocket and poor recognition of the tRNA anticodon have been instrumental in incorporating >200 noncanonical amino acids. PylRS enzymes can be divided into three classes based on their genomic structure. Two classes contain both an N-terminal and C-terminal domain, however the third class (ΔpylSn) lacks the N-terminal domain. In this study we explored the tRNA identity elements for a ΔpylSn tRNAPyl from Candidatus Methanomethylophilus alvus which drives the orthogonality seen with its cognate PylRS (MaPylRS). From aminoacylation and translation assays we identified five key elements in ΔpylSn tRNAPyl necessary for MaPylRS activity. The absence of a base (position 8) and a G-U wobble pair (G28:U42) were found to affect the high-resolution structure of the tRNA, while molecular dynamic simulations led us to acknowledge the rigidity imparted from the G-C base pairs (G3:C70 and G5:C68).
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: