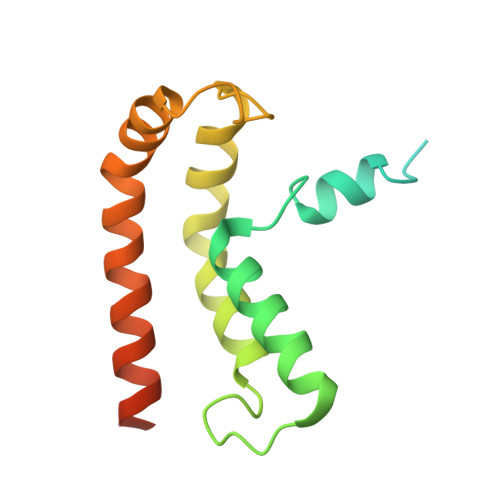
NINJ1 mediates plasma membrane rupture by cutting and releasing membrane disks.
David, L., Borges, J.P., Hollingsworth, L.R., Volchuk, A., Jansen, I., Garlick, E., Steinberg, B.E., Wu, H.(2024) Cell 187: 2224-2235.e16
- PubMed: 38614101
- DOI: https://doi.org/10.1016/j.cell.2024.03.008
- Primary Citation of Related Structures:
8UIP - PubMed Abstract:
The membrane protein NINJ1 mediates plasma membrane rupture in pyroptosis and other lytic cell death pathways. Here, we report the cryo-EM structure of a NINJ1 oligomer segmented from NINJ1 rings. Each NINJ1 subunit comprises amphipathic (⍺1, ⍺2) and transmembrane (TM) helices (⍺3, ⍺4) and forms a chain of subunits, mainly by the TM helices and ⍺1. ⍺3 and ⍺4 are kinked, and the Gly residues are important for function. The NINJ1 oligomer possesses a concave hydrophobic side that should face the membrane and a convex hydrophilic side formed by ⍺1 and ⍺2, presumably upon activation. This structural observation suggests that NINJ1 can form membrane disks, consistent with membrane fragmentation by recombinant NINJ1. Live-cell and super-resolution imaging uncover ring-like structures on the plasma membrane that are released into the culture supernatant. Released NINJ1 encircles a membrane inside, as shown by lipid staining. Therefore, NINJ1-mediated membrane disk formation is different from gasdermin-mediated pore formation, resulting in membrane loss and plasma membrane rupture.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA; Program in Cellular and Molecular Medicine, Boston Children's Hospital, Boston, MA, USA.
Organizational Affiliation: