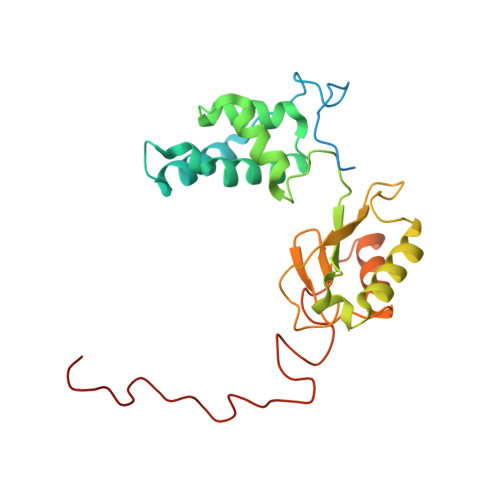
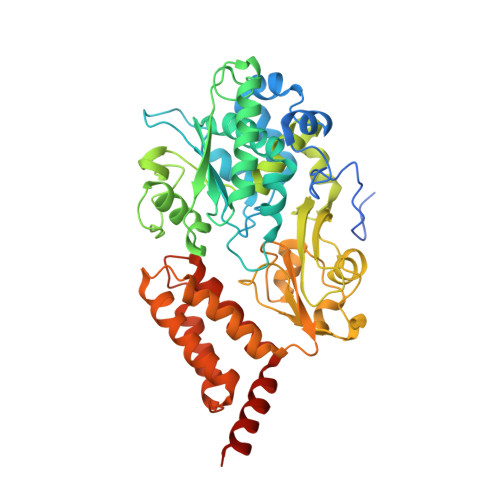
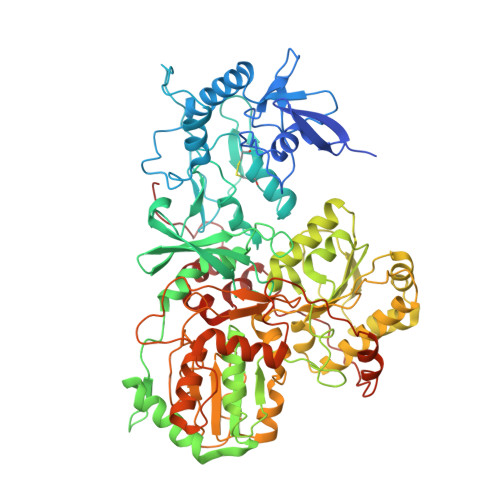
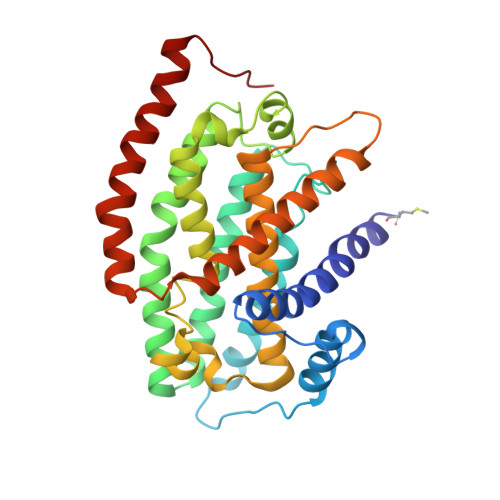
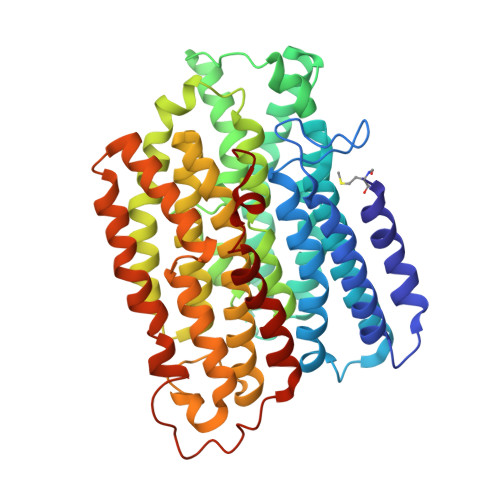
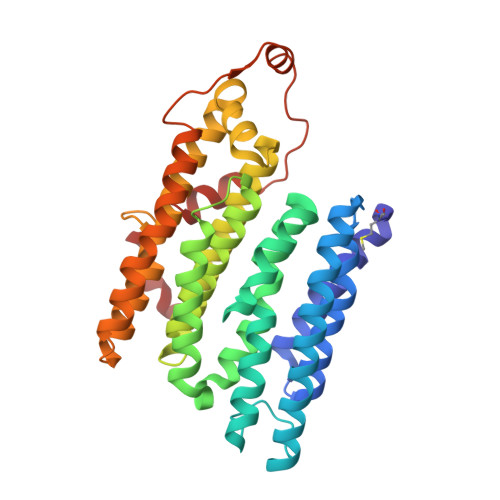
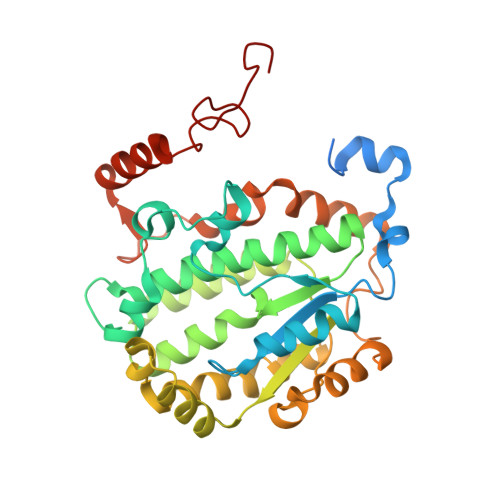
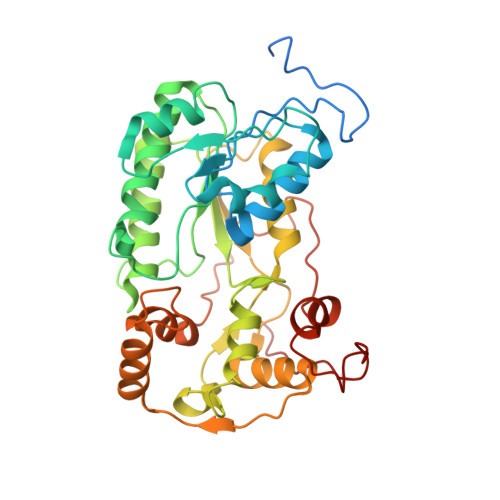
High-resolution in situ structures of mammalian respiratory supercomplexes.
Zheng, W., Chai, P., Zhu, J., Zhang, K.(2024) Nature 631: 232-239
- PubMed: 38811722
- DOI: https://doi.org/10.1038/s41586-024-07488-9
- Primary Citation of Related Structures:
8UD1, 8UEO, 8UEP, 8UEQ, 8UER, 8UES, 8UET, 8UEU, 8UEV, 8UEW, 8UEX, 8UEY, 8UEZ, 8UGD, 8UGE, 8UGF, 8UGG, 8UGH, 8UGI, 8UGJ, 8UGK, 8UGL, 8UGN, 8UGP, 8UGR - PubMed Abstract:
Mitochondria play a pivotal part in ATP energy production through oxidative phosphorylation, which occurs within the inner membrane through a series of respiratory complexes 1-4 . Despite extensive in vitro structural studies, determining the atomic details of their molecular mechanisms in physiological states remains a major challenge, primarily because of loss of the native environment during purification. Here we directly image porcine mitochondria using an in situ cryo-electron microscopy approach. This enables us to determine the structures of various high-order assemblies of respiratory supercomplexes in their native states. We identify four main supercomplex organizations: I 1 III 2 IV 1 , I 1 III 2 IV 2 , I 2 III 2 IV 2 and I 2 III 4 IV 2 , which potentially expand into higher-order arrays on the inner membranes. These diverse supercomplexes are largely formed by 'protein-lipids-protein' interactions, which in turn have a substantial impact on the local geometry of the surrounding membranes. Our in situ structures also capture numerous reactive intermediates within these respiratory supercomplexes, shedding light on the dynamic processes of the ubiquinone/ubiquinol exchange mechanism in complex I and the Q-cycle in complex III. Structural comparison of supercomplexes from mitochondria treated under different conditions indicates a possible correlation between conformational states of complexes I and III, probably in response to environmental changes. By preserving the native membrane environment, our approach enables structural studies of mitochondrial respiratory supercomplexes in reaction at high resolution across multiple scales, from atomic-level details to the broader subcellular context.
- School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China.
Organizational Affiliation: