Characterization and Structural Analyses of Enolase from Shrimp ( Litopenaeus vannamei ).
Chang, X., Zhang, T., Zang, J., Lv, C., Zhao, G.(2023) J Agric Food Chem 71: 19783-19790
- PubMed: 38033172
- DOI: https://doi.org/10.1021/acs.jafc.3c07135
- Primary Citation of Related Structures:
8UEL - PubMed Abstract:
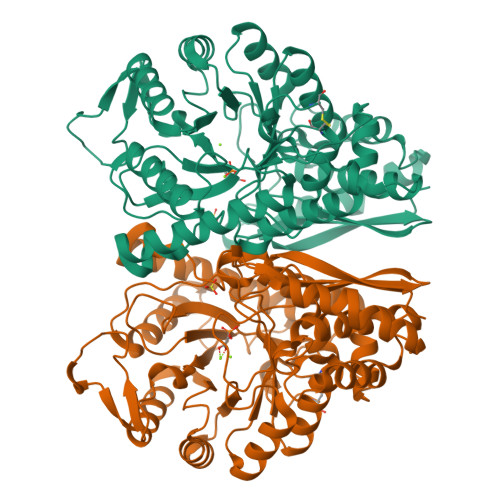
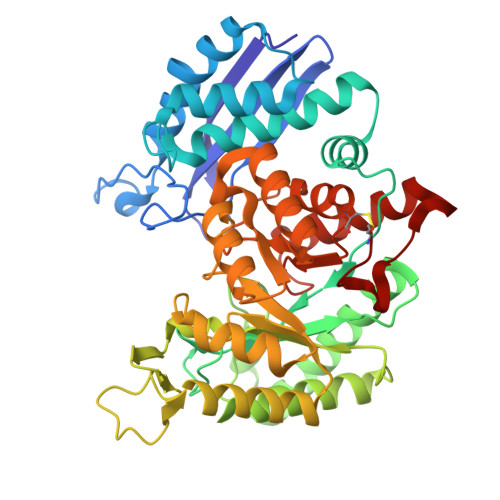
Transcriptome analysis had recognized enolase from shrimp Litopenaeus vannamei ( L. vannamei ), which is termed LvEnolase, as one of the allergens, but its amino acid sequence and protein structure have been lacking. In this study, natural LvEnolase was isolated from L. vannamei and characterized for the first time. The full-length cDNA sequence of LvEnolase was effectively cloned, which encoded 434 amino acid residues. The crystal structure of LvEnolase was successfully determined at a resolution of 2.5 Å by X-ray crystallography (PDB: 8UEL). Notably, it was observed that near the active center, a loop exists in either an open or closed state, and the open loop was associated with the product release phase. Furthermore, enzyme activity assays were conducted to validate the catalytic capabilities of purified LvEnolase. These findings significantly enhance our comprehension of the enolase family and provide valuable support for delving into the functions and characteristics of LvEnolase.
Organizational Affiliation:
College of Food Science & Nutritional Engineering, China Agricultural University, Beijing 100083, China.