Komagataella pastoris Cytochrome c oxidase in complex with human VMAT2 and Reserpine
Ye, J., Liu, B., Li, W.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Synaptic vesicular amine transporter | 514 | Homo sapiens | Mutation(s): 0 Gene Names: SLC18A2 Membrane Entity: Yes | 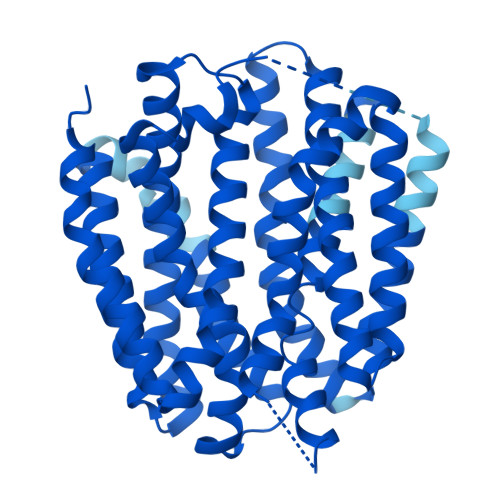 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q05940 (Homo sapiens) Explore Q05940 Go to UniProtKB: Q05940 | |||||
PHAROS: Q05940 GTEx: ENSG00000165646 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q05940 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 1 | B [auth a] | 535 | Komagataella pastoris | Mutation(s): 0 EC: 7.1.1.9 Membrane Entity: Yes | 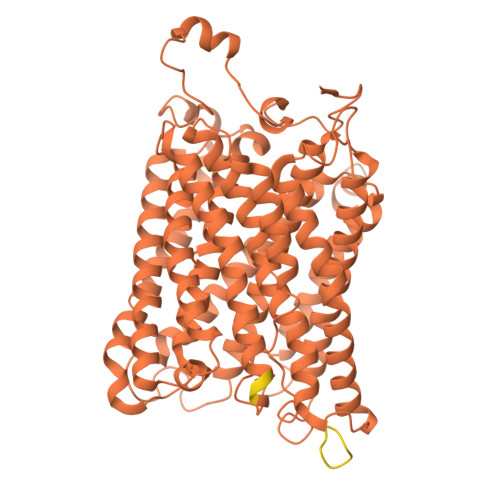 |
UniProt | |||||
Find proteins for F2R0K8 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2R0K8 Go to UniProtKB: F2R0K8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2R0K8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 2 | C [auth b] | 236 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes | 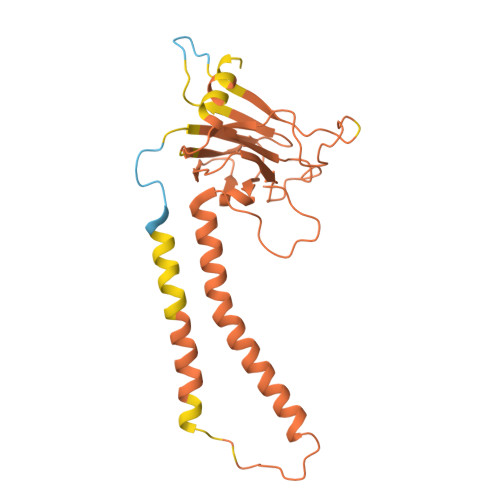 |
UniProt | |||||
Find proteins for F2R0J7 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2R0J7 Go to UniProtKB: F2R0J7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2R0J7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 3 | D [auth c] | 268 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes | 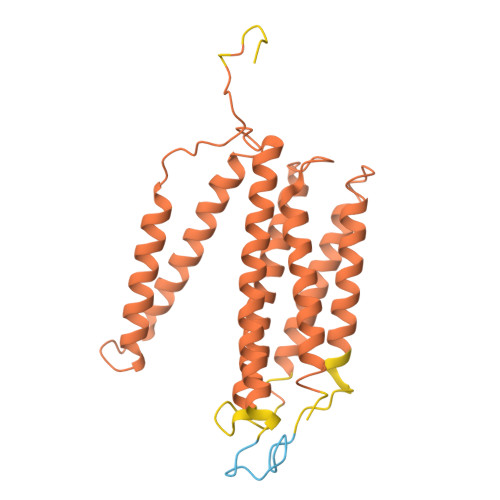 |
UniProt | |||||
Find proteins for F2R0J6 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2R0J6 Go to UniProtKB: F2R0J6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2R0J6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 4 | E [auth d] | 117 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for F2QT92 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2QT92 Go to UniProtKB: F2QT92 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2QT92 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 5 | F [auth e] | 124 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for F2QVW8 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2QVW8 Go to UniProtKB: F2QVW8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2QVW8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 6 | G [auth f] | 100 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for F2QVA2 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2QVA2 Go to UniProtKB: F2QVA2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2QVA2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 7 | H [auth g] | 58 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for F2QS38 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2QS38 Go to UniProtKB: F2QS38 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2QS38 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 8 | I [auth h] | 48 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for F2QRE4 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore F2QRE4 Go to UniProtKB: F2QRE4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F2QRE4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 9 | J [auth i] | 56 | Komagataella pastoris | Mutation(s): 0 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for A0A1G4KPQ9 (Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1)) Explore A0A1G4KPQ9 Go to UniProtKB: A0A1G4KPQ9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1G4KPQ9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEA (Subject of Investigation/LOI) Query on HEA | M [auth a], N [auth a] | HEME-A C49 H56 Fe N4 O6 ZGGYGTCPXNDTRV-PRYGPKJJSA-L |  | ||
| PTY (Subject of Investigation/LOI) Query on PTY | P [auth c], Q [auth c], S [auth e], T [auth i] | PHOSPHATIDYLETHANOLAMINE C40 H80 N O8 P NJGIRBISCGPRPF-KXQOOQHDSA-N |  | ||
| YHR (Subject of Investigation/LOI) Query on YHR | K [auth A] | reserpine C33 H40 N2 O9 QEVHRUUCFGRFIF-MDEJGZGSSA-N |  | ||
| CUA (Subject of Investigation/LOI) Query on CUA | O [auth b] | DINUCLEAR COPPER ION Cu2 ALKZAGKDWUSJED-UHFFFAOYSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | R [auth d] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CU (Subject of Investigation/LOI) Query on CU | L [auth a] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.18.2_3874: |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| American Heart Association | United States | Established Investigator Award |