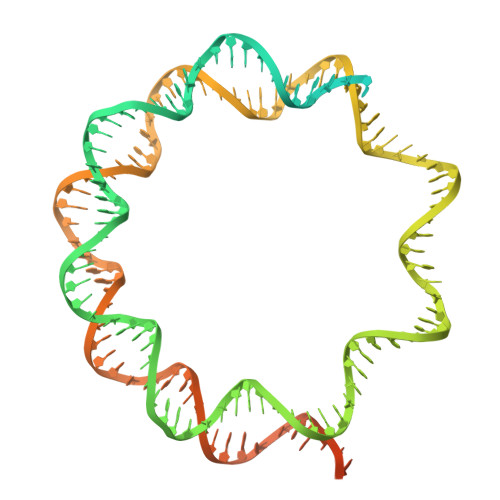
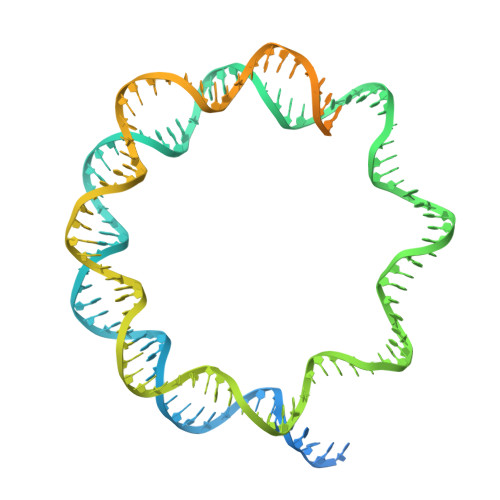
Characterization of Medusavirus encoded histones reveals nucleosome-like structures and a unique linker histone.
Toner, C.M., Hoitsma, N.M., Weerawarana, S., Luger, K.(2024) Nat Commun 15: 9138-9138
- PubMed: 39443461
- DOI: https://doi.org/10.1038/s41467-024-53364-5
- Primary Citation of Related Structures:
8UA7 - PubMed Abstract:
The organization of DNA into nucleosomes is a ubiquitous and ancestral feature that was once thought to be exclusive to the eukaryotic domain of life. Intriguingly, several representatives of the Nucleocytoplasmic Large DNA Viruses (NCLDV) encode histone-like proteins that in Melbournevirus were shown to form nucleosome-like particles. Medusavirus medusae (MM), a distantly related giant virus, encodes all four core histone proteins and, unique amongst most giant viruses, a putative acidic protein with two domains resembling eukaryotic linker histone H1. Here, we report the structure of nucleosomes assembled with MM histones and highlight similarities and differences with eukaryotic and Melbournevirus nucleosomes. Our structure provides insight into how variations in histone tail and loop lengths are accommodated within the context of the nucleosome. We show that MM-histones assemble into tri-nucleosome arrays, and that the putative linker histone H1 does not function in chromatin compaction. These findings expand our limited understanding of chromatin organization by virus-encoded histones.
- Department of Biochemistry, University of Colorado at Boulder, 80309, Boulder, CO, USA.
Organizational Affiliation: